Biogenesis, Trafficking, and Function of Small RNAs in Plants
- PMID: 35251095
- PMCID: PMC8891129
- DOI: 10.3389/fpls.2022.825477
Biogenesis, Trafficking, and Function of Small RNAs in Plants
Abstract
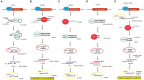
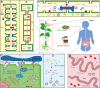
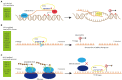
Small RNAs (sRNAs) encoded by plant genomes have received widespread attention because they can affect multiple biological processes. Different sRNAs that are synthesized in plant cells can move throughout the plants, transport to plant pathogens via extracellular vesicles (EVs), and transfer to mammals via food. Small RNAs function at the target sites through DNA methylation, RNA interference, and translational repression. In this article, we reviewed the systematic processes of sRNA biogenesis, trafficking, and the underlying mechanisms of its functions.
Keywords: DNA methylation; RNA interference; biogenesis; functions; small RNA; trafficking; translational repression.
Copyright © 2022 Tang, Yan, Gu and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

