Development and Performance Evaluation of a Novel Ancestry Informative DIP Panel for Continental Origin Inference
- PMID: 35251118
- PMCID: PMC8891605
- DOI: 10.3389/fgene.2021.801275
Development and Performance Evaluation of a Novel Ancestry Informative DIP Panel for Continental Origin Inference
Abstract
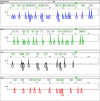
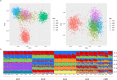
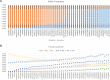
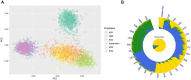
Ancestry informative markers (AIMs) are useful to infer individual biogeographical ancestry and to estimate admixture proportions of admixed populations or individuals. Although a growing number of AIM panels for forensic ancestry origin analyses were developed, they may not efficiently infer the ancestry origins of most populations in China. In this study, a set of 52 ancestry informative deletion/insertion polymorphisms (AIDIPs) were selected with the aim of effectively differentiate continental and partial Chinese populations. All of the selected markers were successfully incorporated into a single multiplex PCR panel, which could be conveniently and efficiently detected on capillary electrophoresis platforms. Genetic distributions of the same 50 AIDIPs in different continental populations revealed that most loci showed high genetic differentiations between East Asian populations and other continental populations. Population genetic analyses of different continental populations indicated that these 50 AIDIPs could clearly discriminate East Asian, European, and African populations. In addition, the 52 AIDIPs also exhibited relatively high cumulative discrimination power in the Eastern Han population, which could be used as a supplementary tool for forensic investigation. Furthermore, the Eastern Han population showed close genetic relationships with East Asian populations and high ancestral components from East Asian populations. In the future, we need to investigate genetic distributions of these 52 AIDIPs in Chinese Han populations in different regions and other ethnic groups, and further evaluate the power of these loci to differentiate different Chinese populations.
Keywords: AIDIP; Eastern Han; ancestry informative marker; deletion/insertion polymorphism; forensic ancestry analysis.
Copyright © 2022 Zhou, Jin, Wu and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor and the reviewer ZW declared as past co‐authorship with one of the authors BZ.
Figures


Similar articles
-
Massively parallel sequencing of 165 ancestry-informative SNPs and forensic biogeographical ancestry inference in three southern Chinese Sinitic/Tai-Kadai populations.Forensic Sci Int Genet. 2021 May;52:102475. doi: 10.1016/j.fsigen.2021.102475. Epub 2021 Feb 2. Forensic Sci Int Genet. 2021. PMID: 33561661
-
A set of novel SNP loci for differentiating continental populations and three Chinese populations.PeerJ. 2019 Mar 29;7:e6508. doi: 10.7717/peerj.6508. eCollection 2019. PeerJ. 2019. PMID: 30956897 Free PMC article.
-
Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing.PLoS One. 2012;7(1):e29684. doi: 10.1371/journal.pone.0029684. Epub 2012 Jan 17. PLoS One. 2012. PMID: 22272242 Free PMC article.
-
Distinguishing three distinct biogeographic regions with an in-house developed 39-AIM-InDel panel and further admixture proportion estimation for Uyghurs.Electrophoresis. 2019 Jun;40(11):1525-1534. doi: 10.1002/elps.201800448. Epub 2019 Mar 5. Electrophoresis. 2019. PMID: 30758063
-
Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean.Ann Hum Genet. 2019 Sep;83(5):348-354. doi: 10.1111/ahg.12320. Epub 2019 Apr 26. Ann Hum Genet. 2019. PMID: 31025319
Cited by
-
Comprehensive evaluations of individual discrimination, kinship analysis, genetic relationship exploration and biogeographic origin prediction in Chinese Dongxiang group by a 60-plex DIP panel.Hereditas. 2023 Mar 29;160(1):14. doi: 10.1186/s41065-023-00271-2. Hereditas. 2023. PMID: 36978173 Free PMC article.
-
Advancements in Modern Nucleic Acid-Based Multiplex Testing Methodologies for the Diagnosis of Swine Infectious Diseases.Vet Sci. 2025 Jul 24;12(8):693. doi: 10.3390/vetsci12080693. Vet Sci. 2025. PMID: 40872643 Free PMC article. Review.
-
Dissecting the genetic admixture and forensic signatures of ethnolinguistically diverse Chinese populations via a 114-plex NGS InDel panel.BMC Genomics. 2024 Nov 25;25(1):1137. doi: 10.1186/s12864-024-10894-y. BMC Genomics. 2024. PMID: 39587470 Free PMC article.
-
Ancestral Information Analysis of Chinese Korean Ethnic Group via a Novel Multiplex DIP System.J Mol Evol. 2023 Dec;91(6):922-934. doi: 10.1007/s00239-023-10143-y. Epub 2023 Nov 25. J Mol Evol. 2023. PMID: 38006428
-
Insertion/deletion polymorphism for genetic background and forensic performance exploration of the Sui group from Guizhou.Heliyon. 2023 Oct 29;9(11):e21384. doi: 10.1016/j.heliyon.2023.e21384. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027767 Free PMC article.
References
LinkOut - more resources
Full Text Sources

