Reconstructing Macroevolutionary Patterns in Avian MHC Architecture With Genomic Data
- PMID: 35251132
- PMCID: PMC8893315
- DOI: 10.3389/fgene.2022.823686
Reconstructing Macroevolutionary Patterns in Avian MHC Architecture With Genomic Data
Abstract
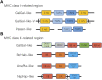
The Major Histocompatibility Complex (MHC) is a hyper-polymorphic genomic region, which forms a part of the vertebrate adaptive immune system and is crucial for intra- and extra-cellular pathogen recognition (MHC-I and MHC-IIA/B, respectively). Although recent advancements in high-throughput sequencing methods sparked research on the MHC in non-model species, the evolutionary history of MHC gene structure is still poorly understood in birds. Here, to explore macroevolutionary patterns in the avian MHC architecture, we retrieved contigs with antigen-presenting MHC and MHC-related genes from available genomes based on third-generation sequencing. We identified: 1) an ancestral avian MHC architecture with compact size and tight linkage between MHC-I, MHC-IIA/IIB and MHC-related genes; 2) three major patterns of MHC-IIA/IIB unit organization in different avian lineages; and 3) lineage-specific gene translocation events (e.g., separation of the antigen-processing TAP genes from the MHC-I region in passerines), and 4) the presence of a single MHC-IIA gene copy in most taxa, showing evidence of strong purifying selection (low dN/dS ratio and low number of positively selected sites). Our study reveals long-term macroevolutionary patterns in the avian MHC architecture and provides the first evidence of important transitions in the genomic arrangement of the MHC region over the last 100 million years of bird evolution.
Keywords: MHC architecture; MHC gene structure; avian MHC; macroevolutionary; third-generation sequencing genome.
Copyright © 2022 He, Liang, Zhu, Dunn, Zhao and Minias.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
MHC architecture in amphibians - ancestral reconstruction, gene rearrangements and duplication patterns.Genome Biol Evol. 2023 May 12;15(5):evad079. doi: 10.1093/gbe/evad079. Online ahead of print. Genome Biol Evol. 2023. PMID: 37170911 Free PMC article.
-
Palaeognaths Reveal Evolutionary Ancestry of the Avian Major Histocompatibility Complex Class II.Genome Biol Evol. 2024 Oct 9;16(10):evae211. doi: 10.1093/gbe/evae211. Genome Biol Evol. 2024. PMID: 39358865 Free PMC article.
-
Ancient duplication, coevolution, and selection at the MHC class IIA and IIB genes of birds.Front Immunol. 2023 Oct 27;14:1250824. doi: 10.3389/fimmu.2023.1250824. eCollection 2023. Front Immunol. 2023. PMID: 37965325 Free PMC article.
-
Avian MHC Evolution in the Era of Genomics: Phase 1.0.Cells. 2019 Sep 26;8(10):1152. doi: 10.3390/cells8101152. Cells. 2019. PMID: 31561531 Free PMC article. Review.
-
Toward an evolutionary genomics of the avian Mhc.Immunol Rev. 1999 Feb;167:119-32. doi: 10.1111/j.1600-065x.1999.tb01386.x. Immunol Rev. 1999. PMID: 10319255 Review.
Cited by
-
Balancing selection shapes population differentiation of major histocompatibility complex genes in wild golden snub-nosed monkeys.Curr Zool. 2023 Sep 24;70(5):596-606. doi: 10.1093/cz/zoad043. eCollection 2024 Oct. Curr Zool. 2023. PMID: 39463695 Free PMC article.
-
Long read genome unravels MHC I genomic architecture, evolution, and diversity loss in Gubernatrix cristata.iScience. 2025 Mar 27;28(5):112301. doi: 10.1016/j.isci.2025.112301. eCollection 2025 May 16. iScience. 2025. PMID: 40491960 Free PMC article.
-
MHC architecture in amphibians - ancestral reconstruction, gene rearrangements and duplication patterns.Genome Biol Evol. 2023 May 12;15(5):evad079. doi: 10.1093/gbe/evad079. Online ahead of print. Genome Biol Evol. 2023. PMID: 37170911 Free PMC article.
-
Some thoughts about what non-mammalian jawed vertebrates are telling us about antigen processing and peptide loading of MHC molecules.Curr Opin Immunol. 2022 Aug;77:102218. doi: 10.1016/j.coi.2022.102218. Epub 2022 Jun 7. Curr Opin Immunol. 2022. PMID: 35687979 Free PMC article. Review.
-
Correlated evolution of oxidative physiology and MHC-based immunosurveillance in birds.Proc Biol Sci. 2024 Jun;291(2025):20240686. doi: 10.1098/rspb.2024.0686. Epub 2024 Jun 19. Proc Biol Sci. 2024. PMID: 38889785 Free PMC article.
References
-
- Biedrzycka A., O’Connor E., Sebastian A., Migalska M., Radwan J., Zając T., et al. (2017). Extreme MHC Class I Diversity in the Sedge Warbler (Acrocephalus Schoenobaenus); Selection Patterns and Allelic Divergence Suggest that Different Genes Have Different Functions. BMC Evol. Biol. 17, 159. 10.1186/s12862-017-0997-9 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

