Targeted Deletion of Centrin in Leishmania braziliensis Using CRISPR-Cas9-Based Editing
- PMID: 35252020
- PMCID: PMC8892584
- DOI: 10.3389/fcimb.2021.790418
Targeted Deletion of Centrin in Leishmania braziliensis Using CRISPR-Cas9-Based Editing
Abstract
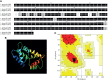
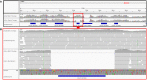
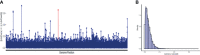
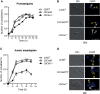
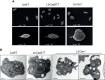
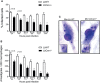
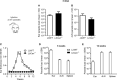
Leishmania braziliensis is the main causative agent of Tegumentary Leishmaniasis in the Americas. However, difficulties related to genome manipulation, experimental infection, and parasite growth have so far limited studies with this species. CRISPR-Cas9-based technology has made genome editing more accessible, and here we have successfully employed the LeishGEdit approach to attenuate L. braziliensis. We generated a transgenic cell line expressing Cas9 and T7 RNA polymerase, which was employed for the targeted deletion of centrin, a calcium-binding cytoskeletal protein involved in the centrosome duplication in eukaryotes. Centrin-deficient Leishmania exhibit growth arrest at the amastigote stage. Whole-genome sequencing of centrin-deficient L. braziliensis (LbCen-/- ) did not indicate the presence of off-target mutations. In vitro, the growth rates of LbCen-/- and wild-type promastigotes were similar, but axenic and intracellular LbCen-/- amastigotes showed a multinucleated phenotype with impaired survival following macrophage infection. Upon inoculation into BALB/c mice, LbCen-/- were detected at an early time point but failed to induce lesion formation, contrary to control animals, infected with wild-type L. braziliensis. A significantly lower parasite burden was also observed in mice inoculated with LbCen-/- , differently from control mice. Given that centrin-deficient Leishmania sp. have become candidates for vaccine development, we propose that LbCen-/- can be further explored for the purposes of immunoprophylaxis against American Tegumentary Leishmaniasis.
Keywords: LeishGEedit; attenuation; genetic manipulation; leishmaniasis; vaccine development.
Copyright © 2022 Sharma, Avendaño Rangel, Reis-Cunha, Marques, Figueira, Borba, Viana, Beneke, Bartholomeu and de Oliveira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The History of Live Attenuated Centrin Gene-Deleted Leishmania Vaccine Candidates.Pathogens. 2022 Apr 2;11(4):431. doi: 10.3390/pathogens11040431. Pathogens. 2022. PMID: 35456106 Free PMC article. Review.
-
Immunization with centrin-Deficient Leishmania braziliensis Does Not Protect against Homologous Challenge.Vaccines (Basel). 2024 Mar 15;12(3):310. doi: 10.3390/vaccines12030310. Vaccines (Basel). 2024. PMID: 38543944 Free PMC article.
-
Effective Genome Editing in Leishmania (Viannia) braziliensis Stably Expressing Cas9 and T7 RNA Polymerase.Front Cell Infect Microbiol. 2021 Nov 10;11:772311. doi: 10.3389/fcimb.2021.772311. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34858879 Free PMC article.
-
Testing of four Leishmania vaccine candidates in a mouse model of infection with Leishmania (Viannia) braziliensis, the main causative agent of cutaneous leishmaniasis in the New World.Clin Vaccine Immunol. 2007 Sep;14(9):1173-81. doi: 10.1128/CVI.00060-07. Epub 2007 Jul 11. Clin Vaccine Immunol. 2007. PMID: 17626159 Free PMC article.
-
Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23.Genes (Basel). 2020 Sep 30;11(10):1159. doi: 10.3390/genes11101159. Genes (Basel). 2020. PMID: 33007987 Free PMC article.
Cited by
-
Draft Genome Sequence of the Protozoan Parasite Leishmania braziliensis Strain BA788, Isolated from a Clinical Case in Bahia State, Brazil.Microbiol Resour Announc. 2022 Dec 15;11(12):e0024522. doi: 10.1128/mra.00245-22. Epub 2022 Nov 1. Microbiol Resour Announc. 2022. PMID: 36318039 Free PMC article.
-
The History of Live Attenuated Centrin Gene-Deleted Leishmania Vaccine Candidates.Pathogens. 2022 Apr 2;11(4):431. doi: 10.3390/pathogens11040431. Pathogens. 2022. PMID: 35456106 Free PMC article. Review.
-
Deletion of MIF gene from live attenuated LdCen-/- parasites enhances protective CD4+ T cell immunity.Sci Rep. 2023 May 5;13(1):7362. doi: 10.1038/s41598-023-34333-2. Sci Rep. 2023. PMID: 37147351 Free PMC article.
-
Next-Generation Leishmanization: Revisiting Molecular Targets for Selecting Genetically Engineered Live-Attenuated Leishmania.Microorganisms. 2023 Apr 16;11(4):1043. doi: 10.3390/microorganisms11041043. Microorganisms. 2023. PMID: 37110466 Free PMC article. Review.
-
Advances in Leishmania Vaccines: Current Development and Future Prospects.Pathogens. 2024 Sep 20;13(9):812. doi: 10.3390/pathogens13090812. Pathogens. 2024. PMID: 39339003 Free PMC article. Review.
References
-
- Andrews S. (2010). FASTQC. A Quality Control Tool for High Throughput Sequence Data, 2010.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

