Autophagy in the Neuronal Ceroid Lipofuscinoses (Batten Disease)
- PMID: 35252181
- PMCID: PMC8888908
- DOI: 10.3389/fcell.2022.812728
Autophagy in the Neuronal Ceroid Lipofuscinoses (Batten Disease)
Abstract
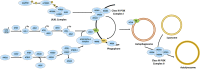
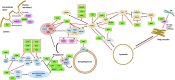
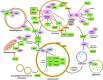
The neuronal ceroid lipofuscinoses (NCLs), also referred to as Batten disease, are a family of neurodegenerative diseases that affect all age groups and ethnicities around the globe. At least a dozen NCL subtypes have been identified that are each linked to a mutation in a distinct ceroid lipofuscinosis neuronal (CLN) gene. Mutations in CLN genes cause the accumulation of autofluorescent lipoprotein aggregates, called ceroid lipofuscin, in neurons and other cell types outside the central nervous system. The mechanisms regulating the accumulation of this material are not entirely known. The CLN genes encode cytosolic, lysosomal, and integral membrane proteins that are associated with a variety of cellular processes, and accumulated evidence suggests they participate in shared or convergent biological pathways. Research across a variety of non-mammalian and mammalian model systems clearly supports an effect of CLN gene mutations on autophagy, suggesting that autophagy plays an essential role in the development and progression of the NCLs. In this review, we summarize research linking the autophagy pathway to the NCLs to guide future work that further elucidates the contribution of altered autophagy to NCL pathology.
Keywords: Batten disease; autophagosome; autophagy; lysosome; mTOR; model system; neurodegeneration; neuronal ceroid lipofucinosis.
Copyright © 2022 Kim, Wilson-Smillie, Thanabalasingam, Lefrancois, Cotman and Huber.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

