Association of SPI1 Haplotypes with Altered SPI1 Gene Expression and Alzheimer's Disease Risk
- PMID: 35253752
- PMCID: PMC9108557
- DOI: 10.3233/JAD-215311
Association of SPI1 Haplotypes with Altered SPI1 Gene Expression and Alzheimer's Disease Risk
Abstract
Background: Genetic studies reveal that single-nucleotide polymorphisms (SNPs) of SPI1 are associated with Alzheimer's disease (AD), while their effects in the Chinese population remain unclear.
Objective: We aimed to examine the AD-association of SPI1 SNPs in the Chinese population and investigate the underlying mechanisms of these SNPs in modulating AD risk.
Methods: We conducted a genetic analysis of three SPI1 SNPs (i.e., rs1057233, rs3740688, and rs78245530) in a Chinese cohort (n = 333 patients with AD, n = 721 normal controls). We also probed public European-descent AD cohorts and gene expression datasets to investigate the putative functions of those SNPs.
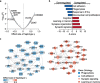
Results: We showed that SPI1 SNP rs3740688 is significantly associated with AD in the Chinese population (odds ratio [OR] = 0.72 [0.58-0.89]) and identified AD-protective SPI1 haplotypes β (tagged by rs1057233 and rs3740688) and γ (tagged by rs3740688 and rs78245530). Specifically, haplotypes β and γ are associated with decreased SPI1 gene expression level in the blood and brain tissues, respectively. The regulatory roles of these haplotypes are potentially mediated by changes in miRNA binding and the epigenetic landscape. Our results suggest that the AD-protective SPI1 haplotypes regulate pathways involved in immune and neuronal functions.
Conclusion: This study is the first to report a significant association of SPI1 with AD in the Chinese population. It also identifies SPI1 haplotypes that are associated with SPI1 gene expression and decreased AD risk.
Keywords: Alzheimer’s disease; SPI1; genetics; haplotype analysis; transcriptome.
Conflict of interest statement
Authors’ disclosures available online (
Figures


References
-
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL (2006) Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63, 168–174. - PubMed
-
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A (1997), Heritability for Alzheimer’s Disease: The Study of Dementia in Swedish Twins. - PubMed
-
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, et al. (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 51, 414–430. - PMC - PubMed
-
- Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, Harari O, Bertelsen S, Fairfax BP, Czajkowski J, Chouraki V, Grenier-Boley B, Bellenguez C, Deming Y, McKenzie A, Raj T, Renton AE, Budde J, Smith A, Fitzpatrick A, Bis JC, DeStefano A, Adams HHH, Ikram MA, Van Der Lee S, Del-Aguila JL, Fernandez MV, Ibañez L, Sims R, Escott-Price V, Mayeux R, Haines JL, Farrer LA, Pericak-Vance MA, Lambert JC, Van Duijn C, Launer L, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Zhang B, Borecki I, Kauwe JSK, Cruchaga C, Hao K, Goate AM (2017) A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci 20, 1052–1061. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

