PCARE requires coiled coil, RP62 kinase-binding and EVH1 domain-binding motifs for ciliary expansion
- PMID: 35253837
- PMCID: PMC9396937
- DOI: 10.1093/hmg/ddac057
PCARE requires coiled coil, RP62 kinase-binding and EVH1 domain-binding motifs for ciliary expansion
Abstract
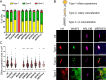
Retinitis pigmentosa (RP) is a genetically heterogeneous form of inherited retinal disease that leads to progressive visual impairment. One genetic subtype of RP, RP54, has been linked to mutations in PCARE (photoreceptor cilium actin regulator). We have recently shown that PCARE recruits WASF3 to the tip of a primary cilium, and thereby activates an Arp2/3 complex which results in the remodeling of actin filaments that drives the expansion of the ciliary tip membrane. On the basis of these findings, and the lack of proper photoreceptor development in mice lacking Pcare, we postulated that PCARE plays an important role in photoreceptor outer segment disk formation. In this study, we aimed to decipher the relationship between predicted structural and function amino acid motifs within PCARE and its function. Our results show that PCARE contains a predicted helical coiled coil domain together with evolutionary conserved binding sites for photoreceptor kinase MAK (type RP62), as well as EVH1 domain-binding linear motifs. Upon deletion of the helical domain, PCARE failed to localize to the cilia. Furthermore, upon deletion of the EVH1 domain-binding motifs separately or together, co-expression of mutant protein with WASF3 resulted in smaller ciliary tip membrane expansions. Finally, inactivation of the lipid modification on the cysteine residue at amino acid position 3 also caused a moderate decrease in the sizes of ciliary tip expansions. Taken together, our data illustrate the importance of amino acid motifs and domains within PCARE in fulfilling its physiological function.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- May-Simera, H., Nagel-Wolfrum, K. and Wolfrum, U. (2017) Cilia – the sensory antennae in the eye. Prog. Retin. Eye Res., 60, 144–180. - PubMed
-
- Roepman, R. and Wolfrum, U. (2007) Protein networks and complexes in photoreceptor cilia. Subcell. Biochem., 43, 209–235. - PubMed
-
- Steinberg, R.H., Fisher, S.K. and Anderson, D.H. (1980) Disc morphogenesis in vertebrate photoreceptors. J. Comp. Neurol., 190, 501–518. - PubMed

