Small Noncoding RNA (sncRNA1) within the Latency-Associated Transcript Modulates Herpes Simplex Virus 1 Virulence and the Host Immune Response during Acute but Not Latent Infection
- PMID: 35254102
- PMCID: PMC9006899
- DOI: 10.1128/jvi.00054-22
Small Noncoding RNA (sncRNA1) within the Latency-Associated Transcript Modulates Herpes Simplex Virus 1 Virulence and the Host Immune Response during Acute but Not Latent Infection
Abstract
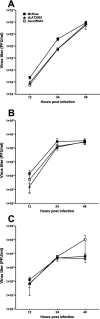
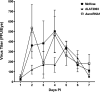
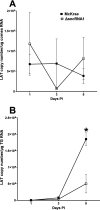
The HSV-1 latency-associated transcript (LAT) locus contains two small noncoding RNA (sncRNA) sequences (sncRNA1 and sncRNA2) that are not microRNAs (miRNAs). We recently reported that sncRNA1 is more important for in vitro activation of the herpesvirus entry mediator than sncRNA2, but its in vivo function is not known. To determine the role, if any, of sncRNA1 during herpes simplex virus 1 (HSV-1) infection in vivo, we deleted the 62-bp sncRNA1 sequence in HSV-1 strain McKrae using dLAT2903 (LAT-minus) virus, creating ΔsncRNA1 recombinant virus. Deletion of the sncRNA1 in ΔsncRNA1 virus was confirmed by complete sequencing of ΔsncRNA1 virus and its parental virus (i.e., McKrae). Replication of ΔsncRNA1 virus in tissue culture or in the eyes of infected mice was similar to that of HSV-1 strain McKrae and dLAT2903 viruses. However, the absence of sncRNA1 significantly reduced the levels of ICP0, ICP4, and gB but not LAT transcripts in infected rabbit skin cells in vitro. In contrast, the absence of sncRNA1 did reduce LAT expression in trigeminal ganglia (TG), but not in corneas, by day 5 postinfection (p.i.) in infected mice. Levels of eye disease in mice infected with ΔsncRNA1 or McKrae virus were similar, and despite reduced LAT levels in TG during acute ΔsncRNA1 infection, McKrae and ΔsncRNA1 viruses did not affect latency or reactivation on day 28 p.i. However, mice infected with ΔsncRNA1 virus were more susceptible to ocular infection than their wild-type (WT) counterparts. Expression of host immune response genes in corneas and TG of infected mice during primary infection showed reduced expression of beta interferon (IFNβ) and IFNγ and altered activation of key innate immune pathways, such as the JAK-STAT pathway in ΔsncRNA1 virus compared with parental WT virus. Our results reveal novel functions for sncRNA1 in upregulating the host immune response and suggest that sncRNA1 has a protective role during primary ocular HSV-1 infection. IMPORTANCE HSV-1 latency-associated transcript (LAT) plays a major role in establishing latency and reactivation; however, the mechanism by which LAT controls these processes is largely unknown. In this study, we sought to establish the role of the small noncoding RNA1 (sncRNA1) encoded within LAT during HSV-1 ocular infection. Our results suggest that sncRNA1 has a protective role during acute ocular infection by modulating the innate immune response to infection.
Keywords: cornea; eye disease; gene expression; immune responses; latency reactivation; recombinant virus; survival; virus replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










Similar articles
-
The anti-apoptotic function of HSV-1 LAT in neuronal cell cultures but not its function during reactivation correlates with expression of two small non-coding RNAs, sncRNA1&2.PLoS Pathog. 2024 Jun 10;20(6):e1012307. doi: 10.1371/journal.ppat.1012307. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38857310 Free PMC article.
-
Herpes Simplex Virus 1 Small Noncoding RNAs 1 and 2 Activate the Herpesvirus Entry Mediator Promoter.J Virol. 2022 Feb 9;96(3):e0198521. doi: 10.1128/JVI.01985-21. Epub 2021 Dec 1. J Virol. 2022. PMID: 34851143 Free PMC article.
-
An M2 Rather than a TH2 Response Contributes to Better Protection against Latency Reactivation following Ocular Infection of Naive Mice with a Recombinant Herpes Simplex Virus 1 Expressing Murine Interleukin-4.J Virol. 2018 Apr 27;92(10):e00051-18. doi: 10.1128/JVI.00051-18. Print 2018 May 15. J Virol. 2018. PMID: 29491152 Free PMC article.
-
The latency-associated gene of herpes simplex virus type 1 (HSV-1) interferes with superinfection by HSV-1.J Neurovirol. 2002 Dec;8 Suppl 2:97-102. doi: 10.1080/13550280290167920. J Neurovirol. 2002. PMID: 12491159 Review.
-
Human alpha-herpesvirus 1 (HSV-1) viral replication and reactivation from latency are expedited by the glucocorticoid receptor.J Virol. 2025 Apr 15;99(4):e0030325. doi: 10.1128/jvi.00303-25. Epub 2025 Mar 27. J Virol. 2025. PMID: 40145740 Free PMC article. Review.
Cited by
-
Multi-targeted loss of the antigen presentation molecule MR1 during HSV-1 and HSV-2 infection.iScience. 2024 Jan 4;27(2):108801. doi: 10.1016/j.isci.2024.108801. eCollection 2024 Feb 16. iScience. 2024. PMID: 38303725 Free PMC article.
-
Glucocorticoid receptor and specificity protein 1 (Sp1) or Sp3, but not the antibiotic Mithramycin A, stimulates human alphaherpesvirus 1 (HSV-1) replication.Antiviral Res. 2024 May;225:105870. doi: 10.1016/j.antiviral.2024.105870. Epub 2024 Mar 29. Antiviral Res. 2024. PMID: 38556059 Free PMC article.
-
The anti-apoptotic function of HSV-1 LAT in neuronal cell cultures but not its function during reactivation correlates with expression of two small non-coding RNAs, sncRNA1&2.PLoS Pathog. 2024 Jun 10;20(6):e1012307. doi: 10.1371/journal.ppat.1012307. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38857310 Free PMC article.
-
A Comparison of Pseudorabies Virus Latency to Other α-Herpesvirinae Subfamily Members.Viruses. 2022 Jun 24;14(7):1386. doi: 10.3390/v14071386. Viruses. 2022. PMID: 35891367 Free PMC article. Review.
References
-
- Mott KR, Bresee CJ, Allen SJ, BenMohamed L, Wechsler SL, Ghiasi H. 2009. Level of herpes simplex virus type 1 latency correlates with severity of corneal scarring and exhaustion of CD8+ T cells in trigeminal ganglia of latently infected mice. J Virol 83:2246–2254. 10.1128/JVI.02234-08. - DOI - PMC - PubMed
-
- Perng GC, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68:8045–8055. 10.1128/jvi.68.12.8045-8055.1994. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

