Fatal affairs - conjugational transfer of a dinoflagellate-killing plasmid between marine Rhodobacterales
- PMID: 35254236
- PMCID: PMC9176285
- DOI: 10.1099/mgen.0.000787
Fatal affairs - conjugational transfer of a dinoflagellate-killing plasmid between marine Rhodobacterales
Abstract
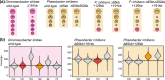
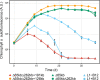
The roseobacter group of marine bacteria is characterized by a mosaic distribution of ecologically important phenotypes. These are often encoded on mobile extrachromosomal replicons. So far, conjugation had only been experimentally proven between the two model organisms Phaeobacter inhibens and Dinoroseobacter shibae. Here, we show that two large natural RepABC-type plasmids from D. shibae can be transferred into representatives of all known major Rhodobacterales lineages. Complete genome sequencing of the newly established Phaeobacter inhibens transconjugants confirmed their genomic integrity. The conjugated plasmids were stably maintained as single copy number replicons in the genuine as well as the new host. Co-cultivation of Phaeobacter inhibens and the transconjugants with the dinoflagellate Prorocentrum minimum demonstrated that Phaeobacter inhibens is a probiotic strain that improves the yield and stability of the dinoflagellate culture. The transconjugant carrying the 191 kb plasmid, but not the 126 kb sister plasmid, killed the dinoflagellate in co-culture.
Keywords: Roseobacter; bacteria–algae interaction; conjugation; horizontal gene transfer.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

