Restriction of viral gene expression and replication prevents immortalization of human keratinocytes by a beta-human papillomavirus
- PMID: 35254896
- PMCID: PMC8931373
- DOI: 10.1073/pnas.2118930119
Restriction of viral gene expression and replication prevents immortalization of human keratinocytes by a beta-human papillomavirus
Abstract
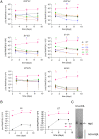
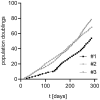
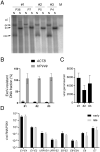
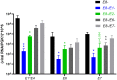
SignificanceHigh-risk (HR) human papillomaviruses (HPV) from the genus alpha cause anogenital and oropharyngeal cancers, whereas the contribution of HPV from the genus beta to the development of cutaneous squamous cell cancer is still under debate. HR-HPV genomes display potent immortalizing activity in human keratinocytes, the natural target cell for HPV. This paper shows that immortalization of keratinocytes by the beta-HPV49 genome requires the inactivation of the viral E8^E2 repressor protein and the presence of the E6 and E7 oncoproteins but also of the E1 and E2 replication proteins. This reveals important differences in the carcinogenic properties of HR-HPV and beta-HPV but also warrants further investigations on the distribution and mutation frequencies of beta-HPV in human cancers.
Keywords: E8^E2; beta-human papillomavirus; cutaneous squamous cell cancer; immortalization.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Bandolin L., et al. , Beta human papillomaviruses infection and skin carcinogenesis. Rev. Med. Virol. 30, e2104 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

