The VIL gene CRAWLING ELEPHANT controls maturation and differentiation in tomato via polycomb silencing
- PMID: 35255095
- PMCID: PMC8939788
- DOI: 10.1371/journal.pgen.1009633
The VIL gene CRAWLING ELEPHANT controls maturation and differentiation in tomato via polycomb silencing
Abstract
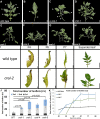
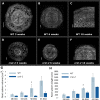
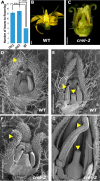
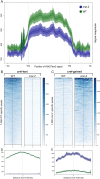
VERNALIZATION INSENSITIVE 3-LIKE (VIL) proteins are PHD-finger proteins that recruit the repressor complex Polycomb Repressive Complex 2 (PRC2) to the promoters of target genes. Most known VIL targets are flowering repressor genes. Here, we show that the tomato VIL gene CRAWLING ELEPHANT (CREL) promotes differentiation throughout plant development by facilitating the trimethylation of Histone H3 on lysine 27 (H3K27me3). We identified the crel mutant in a screen for suppressors of the simple-leaf phenotype of entire (e), a mutant in the AUX/IAA gene ENTIRE/SlIAA9, involved in compound-leaf development in tomato. crel mutants have increased leaf complexity, and suppress the ectopic blade growth of e mutants. In addition, crel mutants are late flowering, and have delayed and aberrant stem, root and flower development. Consistent with a role for CREL in recruiting PRC2, crel mutants show drastically reduced H3K27me3 enrichment at approximately half of the 14,789 sites enriched in wild-type plants, along with upregulation of many underlying genes. Interestingly, this reduction in H3K27me3 across the genome in crel is also associated with gains in H3K27me3 at a smaller number of sites that normally have modest levels of the mark in wild-type plants, suggesting that PRC2 activity is no longer limiting in the absence of CREL. Our results uncover a wide role for CREL in plant and organ differentiation in tomato and suggest that CREL is required for targeting PRC2 activity to, and thus silencing, a specific subset of polycomb targets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
A mutation in the E(Z) methyltransferase that increases trimethylation of histone H3 lysine 27 and causes inappropriate silencing of active Polycomb target genes.Dev Biol. 2012 Apr 15;364(2):249-58. doi: 10.1016/j.ydbio.2011.12.007. Epub 2011 Dec 11. Dev Biol. 2012. PMID: 22182520
-
Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2.Genes (Basel). 2023 Sep 27;14(10):1879. doi: 10.3390/genes14101879. Genes (Basel). 2023. PMID: 37895228 Free PMC article. Review.
-
PHF1 compartmentalizes PRC2 via phase separation.Biochem J. 2023 Nov 29;480(22):1833-1844. doi: 10.1042/BCJ20230040. Biochem J. 2023. PMID: 37888776
-
An ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 is required for proper flowering in Brachypodium distachyon.Plant J. 2018 Mar;93(5):871-882. doi: 10.1111/tpj.13815. Epub 2018 Feb 7. Plant J. 2018. PMID: 29314414
-
A Structural Perspective on Gene Repression by Polycomb Repressive Complex 2.Subcell Biochem. 2021;96:519-562. doi: 10.1007/978-3-030-58971-4_17. Subcell Biochem. 2021. PMID: 33252743 Review.
Cited by
-
Unlocking epigenetic breeding potential in tomato and potato.aBIOTECH. 2024 Oct 23;5(4):507-518. doi: 10.1007/s42994-024-00184-2. eCollection 2024 Dec. aBIOTECH. 2024. PMID: 39650134 Free PMC article. Review.
-
Distinct accessory roles of Arabidopsis VEL proteins in Polycomb silencing.Genes Dev. 2023 Sep 1;37(17-18):801-817. doi: 10.1101/gad.350814.123. Epub 2023 Sep 21. Genes Dev. 2023. PMID: 37734835 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

