UBIAD1 and CoQ10 protect melanoma cells from lipid peroxidation-mediated cell death
- PMID: 35255427
- PMCID: PMC8902599
- DOI: 10.1016/j.redox.2022.102272
UBIAD1 and CoQ10 protect melanoma cells from lipid peroxidation-mediated cell death
Abstract
Cutaneous melanoma is the deadliest type of skin cancer, although it accounts for a minority of all skin cancers. Oxidative stress is involved in all stages of melanomagenesis and cutaneous melanoma can sustain a much higher load of Reactive Oxygen Species (ROS) than normal tissues. Melanoma cells exploit specific antioxidant machinery to support redox homeostasis. The enzyme UBIA prenyltransferase domain-containing protein 1 (UBIAD1) is responsible for the biosynthesis of non-mitochondrial CoQ10 and plays an important role as antioxidant enzyme. Whether UBIAD1 is involved in melanoma progression has not been addressed, yet. Here, we provide evidence that UBIAD1 expression is associated with poor overall survival (OS) in human melanoma patients. Furthermore, UBIAD1 and CoQ10 levels are upregulated in melanoma cells with respect to melanocytes. We show that UBIAD1 and plasma membrane CoQ10 sustain melanoma cell survival and proliferation by preventing lipid peroxidation and cell death. Additionally, we show that the NAD(P)H Quinone Dehydrogenase 1 (NQO1), responsible for the 2-electron reduction of CoQ10 on plasma membranes, acts downstream of UBIAD1 to support melanoma survival. By showing that the CoQ10-producing enzyme UBIAD1 counteracts oxidative stress and lipid peroxidation events in cutaneous melanoma, this work may open to new therapeutic investigations based on UBIAD1/CoQ10 loss to cure melanoma.
Keywords: Antioxidant response; CoQ10; Lipid peroxidation; Melanoma; NQO1; UBIAD1.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
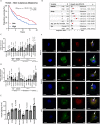

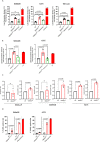
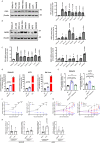
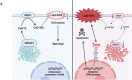
References
-
- Bishal Paudel B., Lewis J.E., Hardeman K.N., Hayford C.E., Robbins C.J., Stauffer P.E., Codreanu S.G., Sherrod S.D., McLean J.A., Kemp M.L., Quatanta V. An integrative gene expression and mathematical flux balance analysis identifies targetable redox vulnerabilities in melanoma cells. Cancer Res. 2021;80:4565–4577. - PMC - PubMed
-
- Dadali T., Diers A.R., Kazerounian S., Muthuswamy S.K., Awate P., Ng R., Mogre S., Spencer C., Krumova K., Rockwell H.E., McDaniel J., Chen E.Y., Gao F., Diedrich K.T., Vemulapalli V., Rodrigues L.O., Akmaev V.R., Thapa K., Hidalgo M., Bose A., Vishnudas V.K., Moser J.A., Granger E., Kiebish M.A., Gesta S., Narain N.R., Sarangarajan R. Elevated levels of mitochondrial CoQ10 induce ROS-mediated apoptosis in pancreatic cancer. Sci. Rep. 2021;11:1–16. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

