The long-term effects of genomic selection: 1. Response to selection, additive genetic variance, and genetic architecture
- PMID: 35255802
- PMCID: PMC8900405
- DOI: 10.1186/s12711-022-00709-7
The long-term effects of genomic selection: 1. Response to selection, additive genetic variance, and genetic architecture
Abstract
Background: Genomic selection has revolutionized genetic improvement in animals and plants, but little is known about its long-term effects. Here, we investigated the long-term effects of genomic selection on response to selection, genetic variance, and the genetic architecture of traits using stochastic simulations. We defined the genetic architecture as the set of causal loci underlying each trait, their allele frequencies, and their statistical additive effects. We simulated a livestock population under 50 generations of phenotypic, pedigree, or genomic selection for a single trait, controlled by either only additive, additive and dominance, or additive, dominance, and epistatic effects. The simulated epistasis was based on yeast data.
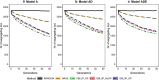
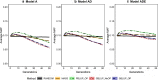
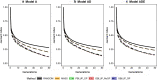
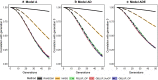
Results: Short-term response was always greatest with genomic selection, while response after 50 generations was greater with phenotypic selection than with genomic selection when epistasis was present, and was always greater than with pedigree selection. This was mainly because loss of genetic variance and of segregating loci was much greater with genomic and pedigree selection than with phenotypic selection. Compared to pedigree selection, selection response was always greater with genomic selection. Pedigree and genomic selection lost a similar amount of genetic variance after 50 generations of selection, but genomic selection maintained more segregating loci, which on average had lower minor allele frequencies than with pedigree selection. Based on this result, genomic selection is expected to better maintain genetic gain after 50 generations than pedigree selection. The amount of change in the genetic architecture of traits was considerable across generations and was similar for genomic and pedigree selection, but slightly less for phenotypic selection. Presence of epistasis resulted in smaller changes in allele frequencies and less fixation of causal loci, but resulted in substantial changes in statistical additive effects across generations.
Conclusions: Our results show that genomic selection outperforms pedigree selection in terms of long-term genetic gain, but results in a similar reduction of genetic variance. The genetic architecture of traits changed considerably across generations, especially under selection and when non-additive effects were present. In conclusion, non-additive effects had a substantial impact on the accuracy of selection and long-term response to selection, especially when selection was accurate.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Hill WG, Kirkpatrick M. What animal breeding has taught us about evolution. Annu Rev Ecol Evol Syst. 2010;41:1–19.
-
- Havenstein GB, Ferket PR, Qureshi MA. Growth, livability, and feed conversion of 1957 versus 2001 broilers when fed representative 1957 and 2001 broiler diets. Poult Sci. 2003;82:1500–1508. - PubMed
-
- Dudley JW, Lambert RJ. 100 generations of selection for oil and protein in corn. Plant Breed Rev. 2003;24:79–110.
-
- Havenstein GB, Ferket PR, Qureshi MA. Carcass composition and yield of 1957 versus 2001 broilers when fed representative 1957 and 2001 broiler diets. Poult Sci. 2003;82:1509–1518. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

