Transcriptomic characterization of the molecular mechanisms induced by RGMa during skeletal muscle nuclei accretion and hypertrophy
- PMID: 35255809
- PMCID: PMC8902710
- DOI: 10.1186/s12864-022-08396-w
Transcriptomic characterization of the molecular mechanisms induced by RGMa during skeletal muscle nuclei accretion and hypertrophy
Abstract
Background: The repulsive guidance molecule a (RGMa) is a GPI-anchor axon guidance molecule first found to play important roles during neuronal development. RGMa expression patterns and signaling pathways via Neogenin and/or as BMP coreceptors indicated that this axon guidance molecule could also be working in other processes and diseases, including during myogenesis. Previous works from our research group have consistently shown that RGMa is expressed in skeletal muscle cells and that its overexpression induces both nuclei accretion and hypertrophy in muscle cell lineages. However, the cellular components and molecular mechanisms induced by RGMa during the differentiation of skeletal muscle cells are poorly understood. In this work, the global transcription expression profile of RGMa-treated C2C12 myoblasts during the differentiation stage, obtained by RNA-seq, were reported.
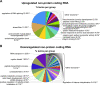
Results: RGMa treatment could modulate the expression pattern of 2,195 transcripts in C2C12 skeletal muscle, with 943 upregulated and 1,252 downregulated. Among them, RGMa interfered with the expression of several RNA types, including categories related to the regulation of RNA splicing and degradation. The data also suggested that nuclei accretion induced by RGMa could be due to their capacity to induce the expression of transcripts related to 'adherens junsctions' and 'extracellular-cell adhesion', while RGMa effects on muscle hypertrophy might be due to (i) the activation of the mTOR-Akt independent axis and (ii) the regulation of the expression of transcripts related to atrophy. Finally, RGMa induced the expression of transcripts that encode skeletal muscle structural proteins, especially from sarcolemma and also those associated with striated muscle cell differentiation.
Conclusions: These results provide comprehensive knowledge of skeletal muscle transcript changes and pathways in response to RGMa.
Keywords: Axon Guidance; Hyperplasia; Hypertrophy; Myogenesis; Skeletal muscle differentiation; Transcriptomic analysis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, et al. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419(6905):392–395. - PubMed
-
- Stahl B, Müller B, von Boxberg Y, Cox EC, Bonhoeffer F. Biochemical characterization of a putative axonal guidance molecule of the chick visual system. Neuron. 1990;5(5):735–743. - PubMed
-
- Müller B, Jay D, Bonhoeffer F. Chromophore-assisted laser inactivation of a repulsive axonal guidance molecule. Curr Biol. 1996;6(11):1497–1502. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

