SnakeCube: containerized and automated pipeline for de novo genome assembly in HPC environments
- PMID: 35255960
- PMCID: PMC8900408
- DOI: 10.1186/s13104-022-05978-5
SnakeCube: containerized and automated pipeline for de novo genome assembly in HPC environments
Abstract
Objective: The rapid progress in sequencing technology and related bioinformatics tools aims at disentangling diversity and conservation issues through genome analyses. The foremost challenges of the field involve coping with questions emerging from the swift development and application of new algorithms, as well as the establishment of standardized analysis approaches that promote transparency and transferability in research.
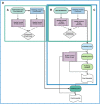
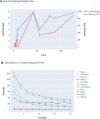
Results: Here, we present SnakeCube, an automated and containerized whole de novo genome assembly pipeline that runs within isolated, secured environments and scales for use in High Performance Computing (HPC) domains. SnakeCube was optimized for its performance and tested for its effectiveness with various inputs, highlighting its safe and robust universal use in the field.
Keywords: Assembly; Container; Genome; Pipeline; de-novo.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources

