Placental uptake and metabolism of 25(OH)vitamin D determine its activity within the fetoplacental unit
- PMID: 35256050
- PMCID: PMC8903835
- DOI: 10.7554/eLife.71094
Placental uptake and metabolism of 25(OH)vitamin D determine its activity within the fetoplacental unit
Abstract
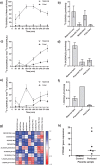
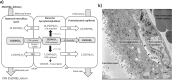
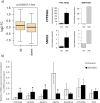
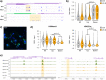
Pregnancy 25-hydroxyvitamin D [25(OH)D] concentrations are associated with maternal and fetal health outcomes. Using physiological human placental perfusion and villous explants, we investigate the role of the placenta in regulating the relationships between maternal 25(OH)D and fetal physiology. We demonstrate active placental uptake of 25(OH)D3 by endocytosis, placental metabolism of 25(OH)D3 into 24,25-dihydroxyvitamin D3 and active 1,25-dihydroxyvitamin D [1,25(OH)2D3], with subsequent release of these metabolites into both the maternal and fetal circulations. Active placental transport of 25(OH)D3 and synthesis of 1,25(OH)2D3 demonstrate that fetal supply is dependent on placental function rather than simply the availability of maternal 25(OH)D3. We demonstrate that 25(OH)D3 exposure induces rapid effects on the placental transcriptome and proteome. These map to multiple pathways central to placental function and thereby fetal development, independent of vitamin D transfer. Our data suggest that the underlying epigenetic landscape helps dictate the transcriptional response to vitamin D treatment. This is the first quantitative study demonstrating vitamin D transfer and metabolism by the human placenta, with widespread effects on the placenta itself. These data demonstrate a complex interplay between vitamin D and the placenta and will inform future interventions using vitamin D to support fetal development and maternal adaptations to pregnancy.
Keywords: Vitamin D; developmental biology; epigenetics; fetal programming; human; placenta; transcriptomics.
© 2022, Ashley et al.
Conflict of interest statement
BA, CS, CJ, FH, JF, FR, EL, EH, LC, PM, RM, SC, HI, JH, KJ, RL, MB, JC No competing interests declared, AM AM is CSO of Proteas Bioanalytics Inc, BioLabs at the Lundquist Institute, CW Cory H White is affiliated with Merck Exploratory Science Centre, Merck Research Laboratories. The author has no financial interests to declare, SB SB part of an academic consortium that has received research funding from Abbott Nutrition, Nestec and Danone, EC EC reports honoraria/travel support from Eli Lilly, UCB, Pfizer and Amgen outside the submitted work, KG KMG has received reimbursement for speaking at conferences sponsored by companies selling nutritional products, and is part of an academic consortium that has received research funding from Abbott Nutrition, Nestec, BenevolentAI Bio Ltd. and Danone, CC CC has received lecture fees and honoraria from Amgen, Danone, Eli Lilly, GSK, Kyowa Kirin, Medtronic, Merck, Nestle, Novartis, Pfizer, Roche, Servier, Shire, Takeda and UCB outside of the submitted work, MH MH has received an honorarium for presenting to Thornton and Ross, SG SG is President and CEO/CTO of Proteas Bioanalytics Inc, BioLabs at the Lundquist Institute, NH NCH reports personal fees, consultancy, lecture fees and honoraria from Alliance for Better Bone Health, AMGEN, MSD, Eli Lilly, Servier, Shire, UCB, Consilient Healthcare, Kyowa Kirin and Internis Pharma, outside the submitted work
Figures



Comment in
-
The extraordinary metabolism of vitamin D.Elife. 2022 Mar 8;11:e77539. doi: 10.7554/eLife.77539. Elife. 2022. PMID: 35257657 Free PMC article.
References
-
- Assar S, Schoenmakers I, Koulman A, Prentice A, Jones KS. UPLC-MS/MS Determination of Deuterated 25-Hydroxyvitamin D (d(3)-25OHD(3)) and Other Vitamin D Metabolites for the Measurement of 25OHD Half-Life, Methods in molecular biology. Methods in Molecular Biology. 2017;2017:257–265. doi: 10.1007/978-1-4939-6730-8_22. - DOI - PubMed
-
- Bikle DD, Gee E, Halloran B, Kowalski MA, Ryzen E, Haddad JG. Assessment of the free fraction of 25-hydroxyvitamin D in serum and its regulation by albumin and the vitamin D-binding protein. The Journal of Clinical Endocrinology and Metabolism. 1986;63:954–959. doi: 10.1210/jcem-63-4-954. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- ISBRC-1215-20014/DH_/Department of Health/United Kingdom
- MC_UP_A620_1015/MRC_/Medical Research Council/United Kingdom
- 17/63/154/DH_/Department of Health/United Kingdom
- 21231/VAC_/Versus Arthritis/United Kingdom
- MC_UU_12011/2/MRC_/Medical Research Council/United Kingdom
- MC_U147585819/MRC_/Medical Research Council/United Kingdom
- MC_PC_21001/MRC_/Medical Research Council/United Kingdom
- U105960371/MRC_/Medical Research Council/United Kingdom
- MC_UP_A620_1014/MRC_/Medical Research Council/United Kingdom
- IS-BRC-1215-20004/DH_/Department of Health/United Kingdom
- BB/P028179/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R21 AI122389/AI/NIAID NIH HHS/United States
- MC_UU_12011/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12011/4/MRC_/Medical Research Council/United Kingdom
- G0400491/MRC_/Medical Research Council/United Kingdom
- RG/15/17/3174/BHF_/British Heart Foundation/United Kingdom
- MC_U147585827/MRC_/Medical Research Council/United Kingdom
- MC_PC_21000/MRC_/Medical Research Council/United Kingdom
- U24 AG047867/AG/NIA NIH HHS/United States
- MC_PC_21003/MRC_/Medical Research Council/United Kingdom
- 4050502589/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 201268/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- G0601019/MRC_/Medical Research Council/United Kingdom
- NF-SI-0515-10042/DH_/Department of Health/United Kingdom
- MC_U147585824/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

