A point-of-care SARS-CoV-2 test based on reverse transcription loop-mediated isothermal amplification without RNA extraction with diagnostic performance same as RT-PCR
- PMID: 35256137
- PMCID: PMC8844505
- DOI: 10.1016/j.aca.2022.339590
A point-of-care SARS-CoV-2 test based on reverse transcription loop-mediated isothermal amplification without RNA extraction with diagnostic performance same as RT-PCR
Abstract
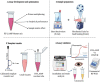
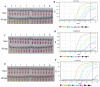
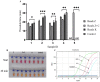
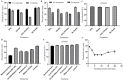
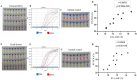
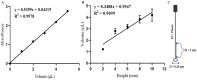
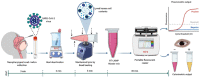
The global public health crisis and economic losses resulting from the current novel coronavirus disease (COVID-19) pandemic have been dire. The most used real-time reverse transcription polymerase chain reaction (RT-PCR) method needs expensive equipment, technical expertise, and a long turnaround time. Therefore, there is a need for a rapid, accurate, and alternative technique of diagnosis that is deployable at resource-poor settings like point-of-care. This study combines heat deactivation and a novel mechanical lysis method by bead beating for quick and simple sample preparation. Then, using an optimized reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay to target genes encoding the open reading frame 8 (ORF8), spike and nucleocapsid proteins of the novel coronavirus, SARS-CoV-2. The test results can be read simultaneously in fluorometric and colorimetric readouts within 40 min from sample collection. We also calibrated a template transfer tool to simplify sample addition into LAMP reactions when pipetting skills are needed. Most importantly, validation of the direct RT-LAMP system based on multiplexing primers S1:ORF8 in a ratio (1:0.8) using 143 patients' nasopharyngeal swab samples showed a diagnostic performance of 99.30% accuracy, with 98.81% sensitivity and 100% selectivity, compared to commercial RT-PCR kits. Since our workflow does not rely on RNA extraction and purification, the time-to-result is two times faster than other workflows with FDA emergency use authorization. Considering all its strengths: speed, simplicity, accuracy and extraction-free, the system can be useful for optimal point-of-care testing of COVID-19.
Keywords: Bead beating; COVID-19; Point-of-care; RT-LAMP; Rapid testing; SARS-CoV-2.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Multisite Clinical Validation of Isothermal Amplification-Based SARS-CoV-2 Detection Assays Using Different Sampling Strategies.Microbiol Spectr. 2021 Oct 31;9(2):e0084621. doi: 10.1128/Spectrum.00846-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668736 Free PMC article.
-
Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA.J Med Microbiol. 2020 Sep;69(9):1169-1178. doi: 10.1099/jmm.0.001238. Epub 2020 Jul 31. J Med Microbiol. 2020. PMID: 32755529 Free PMC article.
-
A Multiplex and Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Sensitive and Rapid Detection of Novel SARS-CoV-2.Front Cell Infect Microbiol. 2021 Jun 29;11:653616. doi: 10.3389/fcimb.2021.653616. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34268131 Free PMC article.
-
Reverse Transcriptase Loop Mediated Isothermal Amplification (RT-LAMP) for COVID-19 diagnosis: a systematic review and meta-analysis.Pathog Glob Health. 2021 Jul;115(5):281-291. doi: 10.1080/20477724.2021.1933335. Epub 2021 Jun 4. Pathog Glob Health. 2021. PMID: 34086539 Free PMC article.
-
Nucleic Acid Testing of SARS-CoV-2.Int J Mol Sci. 2021 Jun 7;22(11):6150. doi: 10.3390/ijms22116150. Int J Mol Sci. 2021. PMID: 34200331 Free PMC article. Review.
Cited by
-
RUNCOV: a one-pot triplex real-time RT-LAMP as a point-of-care diagnostic tool for detecting SARS-CoV-2.Biol Methods Protoc. 2025 Feb 7;10(1):bpaf010. doi: 10.1093/biomethods/bpaf010. eCollection 2025. Biol Methods Protoc. 2025. PMID: 40046730 Free PMC article.
-
Accuracy of reverse-transcription polymerase chain reaction and loop-mediated isothermal amplification in diagnosing severe fever with thrombocytopenia syndrome: A systematic review and meta-analysis.J Med Virol. 2022 Dec;94(12):5922-5932. doi: 10.1002/jmv.28068. Epub 2022 Aug 29. J Med Virol. 2022. PMID: 35968756 Free PMC article.
-
Ultrasensitive and fast detection of SARS-CoV-2 using RT-LAMP without pH-dependent dye.Funct Integr Genomics. 2024 Jan 20;24(1):16. doi: 10.1007/s10142-024-01297-z. Funct Integr Genomics. 2024. PMID: 38242999
-
Development of an Electrochemical Sensor for SARS-CoV-2 Detection Based on Loop-Mediated Isothermal Amplification.Biosensors (Basel). 2023 Oct 11;13(10):924. doi: 10.3390/bios13100924. Biosensors (Basel). 2023. PMID: 37887117 Free PMC article.
-
Characterization of a mouse-adapted strain of bat severe acute respiratory syndrome-related coronavirus.J Virol. 2023 Sep 28;97(9):e0079023. doi: 10.1128/jvi.00790-23. Epub 2023 Aug 21. J Virol. 2023. PMID: 37607058 Free PMC article.
References
-
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

