Monobodies with potent neutralizing activity against SARS-CoV-2 Delta and other variants of concern
- PMID: 35256514
- PMCID: PMC8906176
- DOI: 10.26508/lsa.202101322
Monobodies with potent neutralizing activity against SARS-CoV-2 Delta and other variants of concern
Abstract
Neutralizing antibodies against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are useful for patients' treatment of the coronavirus disease 2019 (COVID-19). We report here affinity maturation of monobodies against the SARS-CoV-2 spike protein and their neutralizing activity against SARS-CoV-2 B.1.1 (Pango v.3.1.14) as well as four variants of concern. We selected matured monobodies from libraries with multi-site saturation mutagenesis on the recognition loops through in vitro selection. One clone, the C4-AM2 monobody, showed extremely high affinity (K D < 0.01 nM) against the receptor-binding domain of the SARS-CoV-2 B.1.1, even in monomer form. Furthermore, the C4-AM2 monobody efficiently neutralized the SARS-CoV-2 B.1.1 (IC 50 = 46 pM, 0.62 ng/ml), and the Alpha (IC 50 = 77 pM, 1.0 ng/ml), Beta (IC 50 = 0.54 nM, 7.2 ng/ml), Gamma (IC 50 = 0.55 nM, 7.4 ng/ml), and Delta (IC 50 = 0.59 nM, 8.0 ng/ml) variants. The obtained monobodies would be useful as neutralizing proteins against current and potentially hazardous future SARS-CoV-2 variants.
© 2022 Kondo et al.
Conflict of interest statement
T Kondo, T Fujino, S Umemoto, G Hayashi, Y Iwatani, and H Murakami are inventors on the provisional patent application (PCT/JP2021/018668; filed 5/18/2021) submitted by Tokai National Higher Education and Research System and National Hospital Organization Nagoya Medical Center. The patent application is for monobody sequences against the SARS-CoV-2 spike protein. Other authors declare that they have no competing interests.
Figures



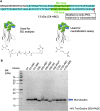


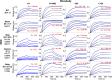



Similar articles
-
Characterization of a Broadly Neutralizing Monoclonal Antibody against SARS-CoV-2 Variants.Viruses. 2022 Jan 24;14(2):230. doi: 10.3390/v14020230. Viruses. 2022. PMID: 35215823 Free PMC article.
-
Sequential Analysis of Binding and Neutralizing Antibody in COVID-19 Convalescent Patients at 14 Months After SARS-CoV-2 Infection.Front Immunol. 2021 Nov 26;12:793953. doi: 10.3389/fimmu.2021.793953. eCollection 2021. Front Immunol. 2021. PMID: 34899762 Free PMC article.
-
Epitope Classification and RBD Binding Properties of Neutralizing Antibodies Against SARS-CoV-2 Variants of Concern.Front Immunol. 2021 Jun 4;12:691715. doi: 10.3389/fimmu.2021.691715. eCollection 2021. Front Immunol. 2021. PMID: 34149735 Free PMC article.
-
Prospects of Neutralizing Nanobodies Against SARS-CoV-2.Front Immunol. 2021 May 28;12:690742. doi: 10.3389/fimmu.2021.690742. eCollection 2021. Front Immunol. 2021. PMID: 34122456 Free PMC article. Review.
-
Monobodies as tool biologics for accelerating target validation and druggable site discovery.RSC Med Chem. 2021 Sep 13;12(11):1839-1853. doi: 10.1039/d1md00188d. eCollection 2021 Nov 17. RSC Med Chem. 2021. PMID: 34820623 Free PMC article. Review.
Cited by
-
Chimeric Protein Switch Biosensors.Adv Biochem Eng Biotechnol. 2024;187:1-35. doi: 10.1007/10_2023_241. Adv Biochem Eng Biotechnol. 2024. PMID: 38273207 Review.
-
ORP9-PH domain-based fluorescent reporters for visualizing phosphatidylinositol 4-phosphate dynamics in living cells.RSC Chem Biol. 2024 Apr 24;5(6):544-555. doi: 10.1039/d3cb00232b. eCollection 2024 Jun 5. RSC Chem Biol. 2024. PMID: 38846081 Free PMC article.
-
Generating a mirror-image monobody targeting MCP-1 via TRAP display and chemical protein synthesis.Nat Commun. 2024 Dec 23;15(1):10723. doi: 10.1038/s41467-024-54902-x. Nat Commun. 2024. PMID: 39715753 Free PMC article.
-
Isolation of an escape-resistant SARS-CoV-2 neutralizing nanobody from a novel synthetic nanobody library.Front Immunol. 2022 Sep 16;13:965446. doi: 10.3389/fimmu.2022.965446. eCollection 2022. Front Immunol. 2022. PMID: 36189235 Free PMC article.
References
-
- Cathcart AL, Havenar-Daughton C, Lempp FA, Ma D, Schmid MA, Agostini ML, Guarino B, Di iulio J, Rosen LE, Tucker H, et al. . (2021) The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. BioRxiv. 10.1101/2021.03.09.434607. (Preprint posted February 18, 2022) - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
