Co-targeting of BAX and BCL-XL proteins broadly overcomes resistance to apoptosis in cancer
- PMID: 35256598
- PMCID: PMC8901805
- DOI: 10.1038/s41467-022-28741-7
Co-targeting of BAX and BCL-XL proteins broadly overcomes resistance to apoptosis in cancer
Abstract
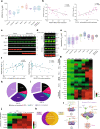
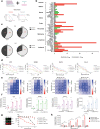
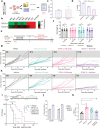
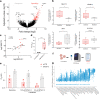
Deregulation of the BCL-2 family interaction network ensures cancer resistance to apoptosis and is a major challenge to current treatments. Cancer cells commonly evade apoptosis through upregulation of the BCL-2 anti-apoptotic proteins; however, more resistant cancers also downregulate or inactivate pro-apoptotic proteins to suppress apoptosis. Here, we find that apoptosis resistance in a diverse panel of solid and hematological malignancies is mediated by both overexpression of BCL-XL and an unprimed apoptotic state, limiting direct and indirect activation mechanisms of pro-apoptotic BAX. Both survival mechanisms can be overcome by the combination of an orally bioavailable BAX activator, BTSA1.2 with Navitoclax. The combination demonstrates synergistic efficacy in apoptosis-resistant cancer cells, xenografts, and patient-derived tumors while sparing healthy tissues. Additionally, functional assays and genomic markers are identified to predict sensitive tumors to the combination treatment. These findings advance the understanding of apoptosis resistance mechanisms and demonstrate a novel therapeutic strategy for cancer treatment.
© 2022. The Author(s).
Conflict of interest statement
E.G., D.E.R., F.K. are inventors on patent applications regarding small molecule BAX activators submitted by Albert Einstein College of Medicine. These patent applications are licensed to BAKX Therapeutics. E.G. is a co-founder and scientific advisor of BAKX Therapeutics, Stelexis Therapeutics, and Selphagy Therapeutics now part of Life Biosciences. E. Vilar has a consulting and advisory role with Janssen Research and Development. None of the above companies have sponsored this research. No potential conflicts of interest were disclosed by the other authors.
Figures



Similar articles
-
Reversal of Mutant KRAS-Mediated Apoptosis Resistance by Concurrent Noxa/Bik Induction and Bcl-2/Bcl-xL Antagonism in Colon Cancer Cells.Mol Cancer Res. 2015 Apr;13(4):659-69. doi: 10.1158/1541-7786.MCR-14-0476. Epub 2014 Dec 29. Mol Cancer Res. 2015. PMID: 25548100 Free PMC article.
-
Targeting the differential addiction to anti-apoptotic BCL-2 family for cancer therapy.Nat Commun. 2017 Jul 17;8:16078. doi: 10.1038/ncomms16078. Nat Commun. 2017. PMID: 28714472 Free PMC article.
-
Combination Treatment with the BRAFV600E Inhibitor Vemurafenib and the BH3 Mimetic Navitoclax for BRAF-Mutant Thyroid Carcinoma.Thyroid. 2019 Apr;29(4):540-548. doi: 10.1089/thy.2018.0511. Thyroid. 2019. PMID: 30869573
-
A brewing understanding of the regulation of Bax function by Bcl-xL and Bcl-2.Mech Ageing Dev. 2017 Jan;161(Pt B):201-210. doi: 10.1016/j.mad.2016.04.007. Epub 2016 Apr 23. Mech Ageing Dev. 2017. PMID: 27112371 Review.
-
Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy.Clin Cancer Res. 2009 Feb 15;15(4):1126-32. doi: 10.1158/1078-0432.CCR-08-0144. Clin Cancer Res. 2009. PMID: 19228717 Free PMC article. Review.
Cited by
-
Antitumor effects of a Sb-rich polyoxometalate on non-small-cell lung cancer by inducing ferroptosis and apoptosis.Chem Sci. 2024 Aug 27;15(37):15367-76. doi: 10.1039/d4sc03856h. Online ahead of print. Chem Sci. 2024. PMID: 39246335 Free PMC article.
-
Mitochondrial Dysfunction-Associated Mechanisms in the Development of Chronic Liver Diseases.Biology (Basel). 2023 Oct 5;12(10):1311. doi: 10.3390/biology12101311. Biology (Basel). 2023. PMID: 37887021 Free PMC article. Review.
-
Bee Pollen as a Source of Biopharmaceuticals for Neurodegeneration and Cancer Research: A Scoping Review and Translational Prospects.Molecules. 2024 Dec 13;29(24):5893. doi: 10.3390/molecules29245893. Molecules. 2024. PMID: 39769981 Free PMC article.
-
PPP2R2A inhibition contributes to preeclampsia by regulating the proliferation, apoptosis, and angiogenesis modulation potential of mesenchymal stem cells.Cell Div. 2024 May 11;19(1):18. doi: 10.1186/s13008-024-00118-w. Cell Div. 2024. PMID: 38734666 Free PMC article.
-
Regulation of apoptosis by ubiquitination in liver cancer.Am J Cancer Res. 2023 Oct 15;13(10):4832-4871. eCollection 2023. Am J Cancer Res. 2023. PMID: 37970337 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

