UCHL5 controls β-catenin destruction complex function through Axin1 regulation
- PMID: 35256667
- PMCID: PMC8901634
- DOI: 10.1038/s41598-022-07642-1
UCHL5 controls β-catenin destruction complex function through Axin1 regulation
Abstract
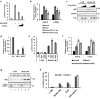
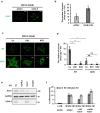
Wnt/β-catenin signaling is crucially involved in many biological processes, from embryogenesis to cancer development. Hence, the complete understanding of its molecular mechanism has been the biggest challenge in the Wnt research field. Here, we identified ubiquitin C-terminal hydrolase like 5 (UCHL5), a deubiquitinating enzyme, as a novel negative regulator of Wnt signaling, upstream of β-catenin. The study further revealed that UCHL5 plays an important role in the β-catenin destruction complex, as it physically interacts with multiple domains of Axin1 protein. Our functional analyses also elucidated that UCHL5 is required for both the stabilization and the polymerization of Axin1 proteins. Interestingly, although these events are governed by deubiquitination in the DIX domain of Axin1 protein, they do not require the deubiquitinating activity of UCHL5. The study proposes a novel molecular mechanism of UCHL5 potentiating the functional activity of Axin1, a scaffolder of the β-catenin destruction complex.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

