Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses
- PMID: 35256775
- PMCID: PMC9305987
- DOI: 10.1038/s41556-022-00853-8
Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses
Abstract
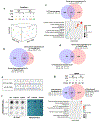
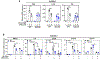
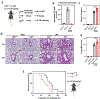
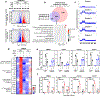
As one of the most induced genes in activated macrophages, immune-responsive gene 1 (IRG1) encodes a mitochondrial metabolic enzyme catalysing the production of itaconic acid (ITA). Although ITA has an anti-inflammatory property, the underlying mechanisms are not fully understood. Here we show that ITA is a potent inhibitor of the TET-family DNA dioxygenases. ITA binds to the same site on TET2 as the co-substrate α-ketoglutarate, inhibiting TET2 catalytic activity. Lipopolysaccharide treatment, which induces Irg1 expression and ITA accumulation, inhibits Tet activity in macrophages. Transcriptome analysis reveals that TET2 is a major target of ITA in suppressing lipopolysaccharide-induced genes, including those regulated by the NF-κB and STAT signalling pathways. In vivo, ITA decreases the levels of 5-hydroxymethylcytosine, reduces lipopolysaccharide-induced acute pulmonary oedema as well as lung and liver injury, and protects mice against lethal endotoxaemia, depending on the catalytic activity of Tet2. Our study thus identifies ITA as an immune modulatory metabolite that selectively inhibits TET enzymes to dampen the inflammatory responses.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

