Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data
- PMID: 35256806
- PMCID: PMC9119698
- DOI: 10.1038/s41588-021-00997-7
Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data
Abstract
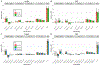
Analyses of data from genome-wide association studies on unrelated individuals have shown that, for human traits and diseases, approximately one-third to two-thirds of heritability is captured by common SNPs. However, it is not known whether the remaining heritability is due to the imperfect tagging of causal variants by common SNPs, in particular whether the causal variants are rare, or whether it is overestimated due to bias in inference from pedigree data. Here we estimated heritability for height and body mass index (BMI) from whole-genome sequence data on 25,465 unrelated individuals of European ancestry. The estimated heritability was 0.68 (standard error 0.10) for height and 0.30 (standard error 0.10) for body mass index. Low minor allele frequency variants in low linkage disequilibrium (LD) with neighboring variants were enriched for heritability, to a greater extent for protein-altering variants, consistent with negative selection. Our results imply that rare variants, in particular those in regions of low linkage disequilibrium, are a major source of the still missing heritability of complex traits and disease.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
The remaining authors declare no competing interests.
Figures
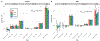

Comment in
-
Discovering missing heritability in whole-genome sequencing data.Nat Genet. 2022 Mar;54(3):224-226. doi: 10.1038/s41588-022-01012-3. Nat Genet. 2022. PMID: 35273401 No abstract available.
References
-
- Lynch M & Walsh B Genetics and analysis of quantitative traits. (Sinauer, 1998).
-
- Fisher RA XV.—The Correlation between Relatives on the Supposition of Mendelian Inheritance. Transactions of the Royal Society of Edinburgh 52, 399–433, doi:10.1017/s0080456800012163 (1918). - DOI
Methods References
Publication types
MeSH terms
Grants and funding
- N01 HC085080/HL/NHLBI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- K01 AG059898/AG/NIA NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- R01 DK110113/DK/NIDDK NIH HHS/United States
- IK6 BX006185/BX/BLRD VA/United States
- R35 HL135818/HL/NHLBI NIH HHS/United States
- K08 HL141601/HL/NHLBI NIH HHS/United States
- R01 HL111314/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- R01 GM075091/GM/NIGMS NIH HHS/United States
- R01 DK124097/DK/NIDDK NIH HHS/United States
- R01 HL158071/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- U54 GM115428/GM/NIGMS NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- R01 MH100141/MH/NIMH NIH HHS/United States
- T32 CA154274/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

