Hibernation slows epigenetic ageing in yellow-bellied marmots
- PMID: 35256811
- PMCID: PMC8986532
- DOI: 10.1038/s41559-022-01679-1
Hibernation slows epigenetic ageing in yellow-bellied marmots
Abstract
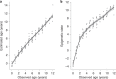
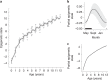
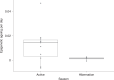
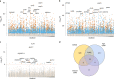
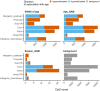
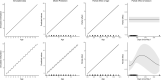
Species that hibernate generally live longer than would be expected based solely on their body size. Hibernation is characterized by long periods of metabolic suppression (torpor) interspersed by short periods of increased metabolism (arousal). The torpor-arousal cycles occur multiple times during hibernation, and it has been suggested that processes controlling the transition between torpor and arousal states cause ageing suppression. Metabolic rate is also a known correlate of longevity; we thus proposed the 'hibernation-ageing hypothesis' whereby ageing is suspended during hibernation. We tested this hypothesis in a well-studied population of yellow-bellied marmots (Marmota flaviventer), which spend 7-8 months per year hibernating. We used two approaches to estimate epigenetic age: the epigenetic clock and the epigenetic pacemaker. Variation in epigenetic age of 149 samples collected throughout the life of 73 females was modelled using generalized additive mixed models (GAMM), where season (cyclic cubic spline) and chronological age (cubic spline) were fixed effects. As expected, the GAMM using epigenetic ages calculated from the epigenetic pacemaker was better able to detect nonlinear patterns in epigenetic ageing over time. We observed a logarithmic curve of epigenetic age with time, where the epigenetic age increased at a higher rate until females reached sexual maturity (two years old). With respect to circannual patterns, the epigenetic age increased during the active season and essentially stalled during the hibernation period. Taken together, our results are consistent with the hibernation-ageing hypothesis and may explain the enhanced longevity in hibernators.
© 2022. The Author(s).
Conflict of interest statement
S.H. is a founder of the non-profit Epigenetic Clock Development Foundation, which plans to license several patents from his employer UC Regents. These patents list S.H. as inventor. The other authors declare no competing interests.
Figures


Comment in
-
The biology of beauty sleep.Nat Ecol Evol. 2022 Apr;6(4):351-352. doi: 10.1038/s41559-022-01683-5. Nat Ecol Evol. 2022. PMID: 35256810 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

