Abrogation of HnRNP L enhances anti-PD-1 therapy efficacy via diminishing PD-L1 and promoting CD8+ T cell-mediated ferroptosis in castration-resistant prostate cancer
- PMID: 35256940
- PMCID: PMC8897216
- DOI: 10.1016/j.apsb.2021.07.016
Abrogation of HnRNP L enhances anti-PD-1 therapy efficacy via diminishing PD-L1 and promoting CD8+ T cell-mediated ferroptosis in castration-resistant prostate cancer
Abstract
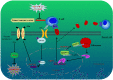
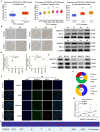
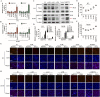
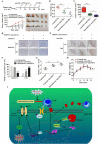
Owing to incurable castration-resistant prostate cancer (CRPC) ultimately developing after treating with androgen deprivation therapy (ADT), it is vital to devise new therapeutic strategies to treat CRPC. Treatments that target programmed cell death protein 1 (PD-1) and programmed death ligand-1 (PD-L1) have been approved for human cancers with clinical benefit. However, many patients, especially prostate cancer, fail to respond to anti-PD-1/PD-L1 treatment, so it is an urgent need to seek a support strategy for improving the traditional PD-1/PD-L1 targeting immunotherapy. In the present study, analyzing the data from our prostate cancer tissue microarray, we found that PD-L1 expression was positively correlated with the expression of heterogeneous nuclear ribonucleoprotein L (HnRNP L). Hence, we further investigated the potential role of HnRNP L on the PD-L1 expression, the sensitivity of cancer cells to T-cell killing and the synergistic effect with anti-PD-1 therapy in CRPC. Indeed, HnRNP L knockdown effectively decreased PD-L1 expression and recovered the sensitivity of cancer cells to T-cell killing in vitro and in vivo, on the contrary, HnRNP L overexpression led to the opposite effect in CRPC cells. In addition, consistent with the previous study, we revealed that ferroptosis played a critical role in T-cell-induced cancer cell death, and HnRNP L promoted the cancer immune escape partly through targeting YY1/PD-L1 axis and inhibiting ferroptosis in CRPC cells. Furthermore, HnRNP L knockdown enhanced antitumor immunity by recruiting infiltrating CD8+ T cells and synergized with anti-PD-1 therapy in CRPC tumors. This study provided biological evidence that HnRNP L knockdown might be a novel therapeutic agent in PD-L1/PD-1 blockade strategy that enhanced anti-tumor immune response in CRPC.
Keywords: ADT, androgen deprivation therapy; Anti-PD-1 therapy; CRPC, castration-resistant prostate cancer; Castration-resistant prostate cancer; DMSO, dimethyl sulfoxide; ELISA, enzyme-linked immunosorbent assay; FBS, fetal bovine serum; Fer-1, ferrostatin-1; Ferroptosis; GSH, glutathione; HnRNP L; HnRNP L, heterogeneous nuclear ribonucleoprotein L; IL, interleukin; INF-γ, interferon gamma; Immune checkpoint blockade; Immune escape; PD-1, programmed cell death protein 1; PD-L1; PD-L1, programmed death ligand1; ROS, reactive oxygen species; STAT, signal transducer and activator of transcription; YY1; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures





References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
-
- Liu J.M., Yu C.P., Chuang H.C., Wu C.T., Hsu R.J. Androgen deprivation therapy for prostate cancer and the risk of autoimmune diseases. Prostate Cancer Prostatic Dis. 2019;22:475–482. - PubMed
-
- Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

