Understanding the physiological functions of the host xenobiotic-sensing nuclear receptors PXR and CAR on the gut microbiome using genetically modified mice
- PMID: 35256948
- PMCID: PMC8897037
- DOI: 10.1016/j.apsb.2021.07.022
Understanding the physiological functions of the host xenobiotic-sensing nuclear receptors PXR and CAR on the gut microbiome using genetically modified mice
Abstract
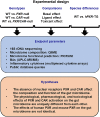
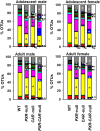
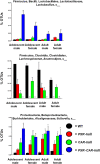
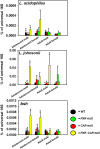
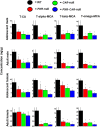
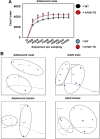
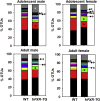
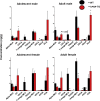
Pharmacological activation of the xenobiotic-sensing nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) is well-known to increase drug metabolism and reduce inflammation. Little is known regarding their physiological functions on the gut microbiome. In this study, we discovered bivalent hormetic functions of PXR/CAR modulating the richness of the gut microbiome using genetically engineered mice. The absence of PXR or CAR increased microbial richness, and absence of both receptors synergistically increased microbial richness. PXR and CAR deficiency increased the pro-inflammatory bacteria Helicobacteraceae and Helicobacter. Deficiency in both PXR and CAR increased the relative abundance of Lactobacillus, which has bile salt hydrolase activity, corresponding to decreased primary taurine-conjugated bile acids (BAs) in feces, which may lead to higher internal burden of taurine and unconjugated BAs, both of which are linked to inflammation, oxidative stress, and cytotoxicity. The basal effect of PXR/CAR on the gut microbiome was distinct from pharmacological and toxicological activation of these receptors. Common PXR/CAR-targeted bacteria were identified, the majority of which were suppressed by these receptors. hPXR-TG mice had a distinct microbial profile as compared to wild-type mice. This study is the first to unveil the basal functions of PXR and CAR on the gut microbiome.
Keywords: BA, bile acid; BSH, bile salt hydrolase; Bile acids; CA, cholic acid; CAR; CAR, constitutive androstane receptor; CDCA, chenodeoxycholic acid; CITCO, 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime; CV, conventional; CYP, cytochrome P450; DCA, deoxycholic acid; EGF, epidermal growth factor; Feces; GF, germ free; GLP-1, glucagon-like peptide-1; GM-CSF, granulocyte-macrophage colony-stimulating factor; Gut microbiome; HDCA, hyodeoxycholic acid; IBD, inflammatory bowel disease; IFNγ, interferon-gamma; IL, interleukin; IS, internal standards; Inflammation; LCA, lithocholic acid; LC–MS/MS, liquid chromatography–tandem mass spectrometry; MCA, muricholic acid; MCP-1, monocyte chemoattractant protein-1; Mice; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NSAID, non-steroidal anti-inflammatory drug; Nuclear receptor; OH, hydroxylated; OTUs, operational taxonomy units; PA, indole-3 propionic acid; PBDEs, polybrominated diphenyl ethers; PCBs, polychlorinated biphenyls; PCoA, Principle Coordinate Analysis; PXR; PXR, pregnane X receptor; PiCRUSt, Phylogenetic Investigation of Communities by Reconstruction of Observed States; QIIME, Quantitative Insights Into Microbial Ecology; SCFAs, short-chain fatty acids; SNP, single-nucleotide polymorphism; SPF, specific-pathogen-free; T, wild type; T-, taurine conjugated; TCPOBOP, 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-Tetrachloro-1,4-bis(pyridyloxy)benzene; TGR-5, Takeda G-protein-coupled receptor 5; TLR4, toll-like receptor 4; TNF, tumor necrosis factor; UDCA, ursodeoxycholic acid; YAP, yes-associated protein; hPXR-TG, humanized PXR transgenic.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures



References
-
- Watanabe M., Houten S.M., Mataki C., Christoffolete M.A., Kim B.W., Sato H., et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

