Targeting a cryptic allosteric site of SIRT6 with small-molecule inhibitors that inhibit the migration of pancreatic cancer cells
- PMID: 35256952
- PMCID: PMC8897208
- DOI: 10.1016/j.apsb.2021.06.015
Targeting a cryptic allosteric site of SIRT6 with small-molecule inhibitors that inhibit the migration of pancreatic cancer cells
Abstract
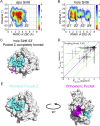
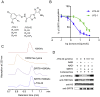
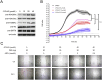
SIRT6 belongs to the conserved NAD+-dependent deacetylase superfamily and mediates multiple biological and pathological processes. Targeting SIRT6 by allosteric modulators represents a novel direction for therapeutics, which can overcome the selectivity problem caused by the structural similarity of orthosteric sites among deacetylases. Here, developing a reversed allosteric strategy AlloReverse, we identified a cryptic allosteric site, Pocket Z, which was only induced by the bi-directional allosteric signal triggered upon orthosteric binding of NAD+. Based on Pocket Z, we discovered an SIRT6 allosteric inhibitor named JYQ-42. JYQ-42 selectively targets SIRT6 among other histone deacetylases and effectively inhibits SIRT6 deacetylation, with an IC50 of 2.33 μmol/L. JYQ-42 significantly suppresses SIRT6-mediated cancer cell migration and pro-inflammatory cytokine production. JYQ-42, to our knowledge, is the most potent and selective allosteric SIRT6 inhibitor. This study provides a novel strategy for allosteric drug design and will help in the challenging development of therapeutic agents that can selectively bind SIRT6.
Keywords: ADPr, ADP-ribose; Allosteric inhibitor; BSA, bull serum albumin; CCK-8, Cell Counting Kit-8; Cell migration; Cytokine production; DMSO, dimethyl sulfoxide; FBS, fetal bovine serum; FDL, Fluor de Lys; H3K18, histone 3 lysine 18; H3K56, histone 3 lysine 56; H3K9, histone 3 lysine 9; HDAC, histone deacetylase; HPLC, high-performance liquid chromatography; IC50, half-maximum inhibitory concentration; IPTG, isopropyl-β-d-thiogalactoside; MD, molecular dynamics; Molecular dynamics simulations; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; PBS, phosphate buffer saline; PMA, phorbol 12-myristate 13-acetate; PMSF, phenylmethanesulfonyl fluoride; Pancreatic cancer; RMSD, root-mean-square deviation; RT-qPCR, real-time quantitative PCR; Reversed allostery; SDS-PAGE, SDS-polyacrylamide gel electrophoresis; SIRT6; SIRT6, sirtuin 6.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures

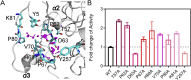


Similar articles
-
Mechanism of allosteric activation of SIRT6 revealed by the action of rationally designed activators.Acta Pharm Sin B. 2021 May;11(5):1355-1361. doi: 10.1016/j.apsb.2020.09.010. Epub 2020 Sep 19. Acta Pharm Sin B. 2021. PMID: 34094839 Free PMC article.
-
Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy.Chem Sci. 2020 Nov 2;12(1):464-476. doi: 10.1039/d0sc05131d. Chem Sci. 2020. PMID: 34163609 Free PMC article.
-
Discovery of a potent and highly selective inhibitor of SIRT6 against pancreatic cancer metastasis in vivo.Acta Pharm Sin B. 2024 Mar;14(3):1302-1316. doi: 10.1016/j.apsb.2023.11.014. Epub 2023 Nov 10. Acta Pharm Sin B. 2024. PMID: 38487000 Free PMC article.
-
The Role of Sirtuin 6 in the Deacetylation of Histone Proteins as a Factor in the Progression of Neoplastic Disease.Int J Mol Sci. 2023 Dec 29;25(1):497. doi: 10.3390/ijms25010497. Int J Mol Sci. 2023. PMID: 38203666 Free PMC article. Review.
-
Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators.J Biol Chem. 2020 Aug 7;295(32):11021-11041. doi: 10.1074/jbc.REV120.011438. Epub 2020 Jun 9. J Biol Chem. 2020. PMID: 32518153 Free PMC article. Review.
Cited by
-
Understanding gilteritinib resistance to FLT3-F691L mutation through an integrated computational strategy.J Mol Model. 2022 Aug 6;28(9):247. doi: 10.1007/s00894-022-05254-0. J Mol Model. 2022. PMID: 35932378
-
Unraveling Small Molecule-Mediated Sirtuin 3 Activation at a Distinct Binding Site for Cardioprotective Therapies.ACS Cent Sci. 2025 Apr 14;11(5):704-718. doi: 10.1021/acscentsci.5c00023. eCollection 2025 May 28. ACS Cent Sci. 2025. PMID: 40454347 Free PMC article.
-
Computer-Aided Drug Design Boosts RAS Inhibitor Discovery.Molecules. 2022 Sep 5;27(17):5710. doi: 10.3390/molecules27175710. Molecules. 2022. PMID: 36080477 Free PMC article. Review.
-
Discovery of a pyrrole-pyridinimidazole derivative as novel SIRT6 inhibitor for sensitizing pancreatic cancer to gemcitabine.Cell Death Dis. 2023 Aug 4;14(8):499. doi: 10.1038/s41419-023-06018-1. Cell Death Dis. 2023. PMID: 37542062 Free PMC article.
-
Mechanistic basis of N-terminal domain-mediated allostery in SIRT6: integrating molecular dynamics simulations and biochemical assays.Mol Divers. 2025 Aug 30. doi: 10.1007/s11030-025-11340-1. Online ahead of print. Mol Divers. 2025. PMID: 40885886
References
LinkOut - more resources
Full Text Sources

