RADx Variant Task Force Program for Assessing the Impact of Variants on SARS-CoV-2 Molecular and Antigen Tests
- PMID: 35257097
- PMCID: PMC8864940
- DOI: 10.1109/OJEMB.2021.3116490
RADx Variant Task Force Program for Assessing the Impact of Variants on SARS-CoV-2 Molecular and Antigen Tests
Abstract
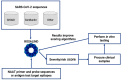
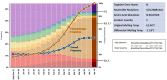
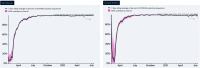
Goal: Monitoring the genetic diversity and emerging mutations of SARS-CoV-2 is crucial for understanding the evolution of the virus and assuring the performance of diagnostic tests, vaccines, and therapies against COVID-19. SARS-CoV-2 is still adapting to humans and, as illustrated by B.1.1.7 (Alpha) and B.1.617.2 (Delta), lineage dynamics are fluid, and strain prevalence may change radically in a matter of months. The National Institutes of Health's Rapid Acceleration of Diagnostics (RADxSM) initiative created a Variant Task Force to assess the impact of emerging SARS-CoV-2 variants on in vitro diagnostic testing. Working in tandem with clinical laboratories, the FDA, and the CDC, the Variant Task Force uses both in silico modeling and in vitro testing to determine the effect of SARS-CoV-2 mutations on diagnostic molecular and antigen tests. Here, we offer an overview of the approach and activities of the RADx Variant Task Force to ensure test performance against emerging SARS-CoV-2 lineages.
Keywords: COVID-19; SARS-CoV-2; in vitro diagnostics; mutations; variants of concern.
Figures
References
-
- Zheng J., “SARS-CoV-2: An emerging coronavirus that causes a global threat,” Int. J. Biol. Sci., vol. 16, no. 10, pp. 1678–1685, Mar. 2020. [Online]. Available: https://www.ijbs.com/v16p1678.htm - PMC - PubMed
-
- Center for Systems Science and Engineering at Johns Hopkins University, COVID-19 Dashboard, Johns Hopkins University & Medicine, USA, Oct. 2021. [Online]. Available: https://coronavirus.jhu.edu/map.html
-
- Zhou P. et al., “A pneumonia outbreak associated with a new coronavirus of probable bat origin,” Nature, vol. 579, no. 7798, pp. 270–273, Feb. 2020, [Online]. Available: https://www.nature.com/articles/s41586-020-2012-7 - PMC - PubMed
-
- Jo W. et al., “The evolutionary dynamics of endemic human coronaviruses,” Virus Evol., vol. 7, no. 1, pp. 1–12, Jan. 2021, [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7980080/ - PMC - PubMed
-
- Callaway E., “Making sense of coronavirus mutations,” Nature, vol. 585, no. 7824, pp. 174–177, Sep. 2020, [Online]. Available: https://media.nature.com/original/magazine-assets/d41586-020-02544-6/d41... - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

