Revisiting SARS-CoV-2 environmental contamination by patients with COVID-19: The Omicron variant does not differ from previous strains
- PMID: 35257907
- PMCID: PMC8896873
- DOI: 10.1016/j.ijid.2022.03.001
Revisiting SARS-CoV-2 environmental contamination by patients with COVID-19: The Omicron variant does not differ from previous strains
Abstract
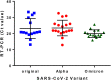
SARS-CoV-2 Omicron strain emergence raised concerns that its enhanced infectivity is partly due to altered spread/contamination modalities. We therefore sampled high-contact surfaces and air in close proximity to patients who were verified as infected with the Omicron strain, using identical protocols applied to sample patients positive to the original or Alpha strains. Cumulatively, for all 3 strains, viral RNA was detected in 90 of 168 surfaces and 6 of 49 air samples (mean cycle threshold [Ct]=35.2±2.5). No infective virus was identified. No significant differences in prevalence were found between strains.
Keywords: Alpha; Omicron; SARS-CoV-2; air samples; original; surface contamination.
Copyright © 2022 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Conflict of interest None declared.
Figures
Similar articles
-
Clinical and Pulmonary CT Characteristics of Patients Infected With the SARS-CoV-2 Omicron Variant Compared With Those of Patients Infected With the Alpha Viral Strain.Front Public Health. 2022 Jul 12;10:931480. doi: 10.3389/fpubh.2022.931480. eCollection 2022. Front Public Health. 2022. PMID: 35903393 Free PMC article.
-
SARS-CoV-2 Omicron Variant, Lineage BA.1, Is Associated with Lower Viral Load in Nasopharyngeal Samples Compared to Delta Variant.Viruses. 2022 Apr 28;14(5):919. doi: 10.3390/v14050919. Viruses. 2022. PMID: 35632661 Free PMC article.
-
[Comparative study of Wuhan-like and omicron-like variants of SARS-CoV-2 in experimental animal models].Vopr Virusol. 2022 Nov 19;67(5):439-449. doi: 10.36233/0507-4088-135. Vopr Virusol. 2022. PMID: 36515289 Russian.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
SARS-CoV-2 Omicron variant: recent progress and future perspectives.Signal Transduct Target Ther. 2022 Apr 28;7(1):141. doi: 10.1038/s41392-022-00997-x. Signal Transduct Target Ther. 2022. PMID: 35484110 Free PMC article. Review.
Cited by
-
Environmental Contamination of SARS-CoV-2 Delta VOC by COVID-19 Patients Staying in the Hospital for More Than Two Weeks.Risk Manag Healthc Policy. 2023 Oct 17;16:2163-2170. doi: 10.2147/RMHP.S413639. eCollection 2023. Risk Manag Healthc Policy. 2023. PMID: 37868023 Free PMC article.
-
SARS-CoV-2 indoor environment contamination with epidemiological and experimental investigations.Indoor Air. 2022 Oct;32(10):e13118. doi: 10.1111/ina.13118. Indoor Air. 2022. PMID: 36305066 Free PMC article.
-
Negligible risk of surface transmission of SARS-CoV-2 in public transportation.J Travel Med. 2023 Sep 5;30(5):taad065. doi: 10.1093/jtm/taad065. J Travel Med. 2023. PMID: 37133444 Free PMC article.
-
Assessment of environmental surface contamination with SARS-CoV-2 in concert halls and banquet rooms in Japan.J Infect Chemother. 2023 Jun;29(6):604-609. doi: 10.1016/j.jiac.2023.02.013. Epub 2023 Mar 7. J Infect Chemother. 2023. PMID: 36894016 Free PMC article.
-
Development, testing and validation of a SARS-CoV-2 multiplex panel for detection of the five major variants of concern on a portable PCR platform.Front Public Health. 2022 Dec 15;10:1042647. doi: 10.3389/fpubh.2022.1042647. eCollection 2022. Front Public Health. 2022. PMID: 36590003 Free PMC article.
References
-
- Ben-Shmuel A, Brosh-Nissimov T, Glinert I, Bar-David E, Sittner A, Poni R, et al. Detection and infectivity potential of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clinical Microbiology and Infection. 2020;26:1658–16662. - PMC - PubMed
-
- Kannan S, Shaik P, Ali S, Sheeza A. Omicron (B.1.1.529)- variant of concern- molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci. 2021;25:8019–8022. - PubMed
-
- Lane M, Brownsword E, Babiker A, Ingersoll J, Waggoner J, Ayers M, et al. Bioaerosol sampling for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in referral center withcritical ill coronavirus disease 2019 (COVID-19) patients march-may 2020. Clinical Infectious Diseases. 2021;73:1790–1794. - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

