Digital PCR Applications in the SARS-CoV-2/COVID-19 Era: a Roadmap for Future Outbreaks
- PMID: 35258315
- PMCID: PMC9491181
- DOI: 10.1128/cmr.00168-21
Digital PCR Applications in the SARS-CoV-2/COVID-19 Era: a Roadmap for Future Outbreaks
Erratum in
-
Erratum for Nyaruaba et al., "Digital PCR Applications in the SARS-CoV-2/COVID-19 Era: a Roadmap for Future Outbreaks".Clin Microbiol Rev. 2023 Jun 21;36(2):e0005223. doi: 10.1128/cmr.00052-23. Epub 2023 May 30. Clin Microbiol Rev. 2023. PMID: 37249467 Free PMC article. No abstract available.
Abstract
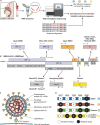
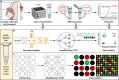
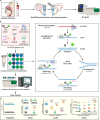
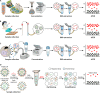
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to a global public health disaster. The current gold standard for the diagnosis of infected patients is real-time reverse transcription-quantitative PCR (RT-qPCR). As effective as this method may be, it is subject to false-negative and -positive results, affecting its precision, especially for the detection of low viral loads in samples. In contrast, digital PCR (dPCR), the third generation of PCR, has been shown to be more effective than the gold standard, RT-qPCR, in detecting low viral loads in samples. In this review article, we selected publications to show the broad-spectrum applications of dPCR, including the development of assays and reference standards, environmental monitoring, mutation detection, and clinical diagnosis of SARS-CoV-2, while comparing it analytically to the gold standard, RT-qPCR. In summary, it is evident that the specificity, sensitivity, reproducibility, and detection limits of RT-dPCR are generally unaffected by common factors that may affect RT-qPCR. As this is the first time that dPCR is being tested in an outbreak of such a magnitude, knowledge of its applications will help chart a course for future diagnosis and monitoring of infectious disease outbreaks.
Keywords: COVID-19; RT-dPCR; RT-qPCR; SARS-CoV-2; ddPCR; diagnosis; quantification; viral load.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR.Talanta. 2021 Mar 1;224:121726. doi: 10.1016/j.talanta.2020.121726. Epub 2020 Oct 27. Talanta. 2021. PMID: 33379001 Free PMC article.
-
Digital PCR assay for the effective detection of COVID-19 patients with SARS-CoV-2 low viral load.J Virol Methods. 2021 Sep;295:114185. doi: 10.1016/j.jviromet.2021.114185. Epub 2021 May 26. J Virol Methods. 2021. PMID: 34051244 Free PMC article.
-
Droplet digital PCR application for the detection of SARS-CoV-2 in air sample.Front Public Health. 2023 Oct 27;11:1208348. doi: 10.3389/fpubh.2023.1208348. eCollection 2023. Front Public Health. 2023. PMID: 37965510 Free PMC article.
-
Digital PCR in Virology: Current Applications and Future Perspectives.Mol Diagn Ther. 2025 Jan;29(1):43-54. doi: 10.1007/s40291-024-00751-9. Epub 2024 Nov 2. Mol Diagn Ther. 2025. PMID: 39487879 Review.
-
RT-qPCR Testing and Performance Metrics in the COVID-19 Era.Int J Mol Sci. 2024 Aug 28;25(17):9326. doi: 10.3390/ijms25179326. Int J Mol Sci. 2024. PMID: 39273275 Free PMC article. Review.
Cited by
-
A culture-free method for rapidly and accurately quantifying active SARS-CoV-2.Anal Bioanal Chem. 2023 Sep;415(23):5745-5753. doi: 10.1007/s00216-023-04855-9. Epub 2023 Jul 24. Anal Bioanal Chem. 2023. PMID: 37486370
-
Development and validation of nanoplate-based RT-dPCR assay for canine respiratory coronavirus detection in various clinical samples.BMC Vet Res. 2025 May 17;21(1):350. doi: 10.1186/s12917-025-04807-8. BMC Vet Res. 2025. PMID: 40382640 Free PMC article.
-
The first detection of SARS-CoV-2 RNA in the wastewater of Bucharest, Romania.Sci Rep. 2024 Sep 17;14(1):21730. doi: 10.1038/s41598-024-72854-6. Sci Rep. 2024. PMID: 39289536 Free PMC article.
-
Rapid and accurate detection of SARS-CoV-2 using the RHAM technology.Sci Rep. 2023 Dec 20;13(1):22798. doi: 10.1038/s41598-023-49733-7. Sci Rep. 2023. PMID: 38129524 Free PMC article.
-
Tackling Infectious Diseases with Rapid Molecular Diagnosis and Innovative Prevention.Infect Dis Rep. 2024 Mar 5;16(2):216-227. doi: 10.3390/idr16020017. Infect Dis Rep. 2024. PMID: 38525764 Free PMC article. Review.
References
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Chatterjee N, Mahmood Z, Marcussen E. 2021. Politics of vaccine nationalism in India: global and domestic implications. Forum Dev Stud 48:357–369. 10.1080/08039410.2021.1918238. - DOI
-
- Lucas C, Vogels CBF, Yildirim I, Rothman JE, Lu P, Monteiro V, Gelhausen JR, Campbell M, Silva J, Tabachikova A, Peña-Hernandez MA, Muenker MC, Breban MI, Fauver JR, Mohanty S, Huang J, Yale SARS-CoV-2 Genomic Surveillance Initiative, Shaw AC, Ko AI, Omer SB, Grubaugh ND, Iwasaki A. 2021. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature 600:523–529. 10.1038/s41586-021-04085-y. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

