Inhibitors of PARP: Number crunching and structure gazing
- PMID: 35259019
- PMCID: PMC8931346
- DOI: 10.1073/pnas.2121979119
Inhibitors of PARP: Number crunching and structure gazing
Abstract
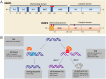
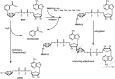
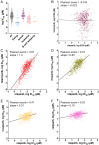
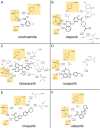
SignificancePARP is an important target in the treatment of cancers, particularly in patients with breast, ovarian, or prostate cancer that have compromised homologous recombination repair (i.e., BRCA-/-). This review about inhibitors of PARP (PARPi) is for readers interested in the development of next-generation drugs for the treatment of cancer, providing insights into structure-activity relationships, in vitro vs. in vivo potency, PARP trapping, and synthetic lethality.
Keywords: HPF1; cancer drugs; drug specificity; inhibitor of Parp; synthetic lethality.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Lodish H., et al. , Molecular Biology of the Cell (Freeman, New York, NY, 2016).
-
- Jeggo P. A., Pearl L. H., Carr A. M., DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 16, 35–42 (2016). - PubMed
-
- Wood R. D., Mitchell M., Sgouras J., Lindahl T., Human DNA repair genes. Science 291, 1284–1289 (2001). - PubMed
-
- Gibson B. A., Kraus W. L., New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012). - PubMed
-
- Bai P., Biology of poly(ADP-ribose) polymerases: The factotums of cell maintenance. Mol. Cell 58, 947–958 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

