Biological mechanisms of aging predict age-related disease co-occurrence in patients
- PMID: 35259281
- PMCID: PMC9009120
- DOI: 10.1111/acel.13524
Biological mechanisms of aging predict age-related disease co-occurrence in patients
Abstract
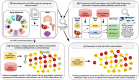
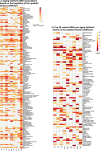
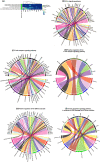
Genetic, environmental, and pharmacological interventions into the aging process can confer resistance to multiple age-related diseases in laboratory animals, including rhesus monkeys. These findings imply that individual mechanisms of aging might contribute to the co-occurrence of age-related diseases in humans and could be targeted to prevent these conditions simultaneously. To address this question, we text mined 917,645 literature abstracts followed by manual curation and found strong, non-random associations between age-related diseases and aging mechanisms in humans, confirmed by gene set enrichment analysis of GWAS data. Integration of these associations with clinical data from 3.01 million patients showed that age-related diseases associated with each of five aging mechanisms were more likely than chance to be present together in patients. Genetic evidence revealed that innate and adaptive immunity, the intrinsic apoptotic signaling pathway and activity of the ERK1/2 pathway were associated with multiple aging mechanisms and diverse age-related diseases. Mechanisms of aging hence contribute both together and individually to age-related disease co-occurrence in humans and could potentially be targeted accordingly to prevent multimorbidity.
Keywords: age-related disease; aging; aging hallmarks; genetics; multimorbidity.
© 2022 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no financial conflicts of interest to disclose. At the time of conducting this research, MZ was employed at BenevolentAI. Since completing the work, MZ is now a full‐time employee of GlaxoSmithKline.
Figures


References
-
- Amell, A. , Roso‐Llorach, A. , Palomero, L. , Cuadras, D. , Galván‐Femenía, I. , Serra‐Musach, J. , Comellas, F. , de Cid, R. , Pujana, M. A. , & Violán, C. (2018). Disease networks identify specific conditions and pleiotropy influencing multimorbidity in the general population. Scientific Reports, 8, 15970. 10.1038/s41598-018-34361-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

