Robust regulatory architecture of pan-neuronal gene expression
- PMID: 35259341
- PMCID: PMC9050922
- DOI: 10.1016/j.cub.2022.02.040
Robust regulatory architecture of pan-neuronal gene expression
Abstract
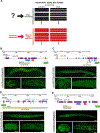
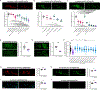
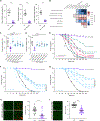
Pan-neuronally expressed genes, such as genes involved in the synaptic vesicle cycle or in neuropeptide maturation, are critical for proper function of all neurons, but the transcriptional control mechanisms that direct such genes to all neurons of a nervous system remain poorly understood. We show here that six members of the CUT family of homeobox genes control pan-neuronal identity specification in Caenorhabditis elegans. Single CUT mutants show barely any effects on pan-neuronal gene expression or global nervous system function, but such effects become apparent and progressively worsen upon removal of additional CUT family members, indicating a critical role of gene dosage. Overexpression of each individual CUT gene rescued the phenotype of compound mutants, corroborating that gene dosage, rather than the activity of specific members of the gene family, is critical for CUT gene family function. Genome-wide binding profiles, as well as mutation of CUT homeodomain binding sites by CRISPR/Cas9 genome engineering show that CUT genes directly control the expression of pan-neuronal features. Moreover, CUT genes act in conjunction with neuron-type-specific transcription factors to control pan-neuronal gene expression. Our study, therefore, provides a previously missing key insight into how neuronal gene expression programs are specified and reveals a highly buffered and robust mechanism that controls the most critical functional features of all neuronal cell types.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

