Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes
- PMID: 35259661
- PMCID: PMC8881819
- DOI: 10.1016/j.compbiolchem.2022.107657
Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes
Abstract
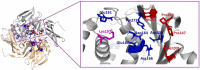
The novel coronavirus disease (COVID-19) is a highly contagious disease caused by the SARS-CoV-2 virus, leading severe acute respiratory syndrome in patients. Although various antiviral drugs and their combinations have been tried so far against SARS-CoV-2 and they have shown some effectiveness, there is still a need for safe and cost-effective binding inhibitors in the fight against COVID-19. Therefore, phytochemicals in nature can be a quick solution due to their wide therapeutic spectrum and strong antiviral, anti-inflammatory, and antioxidant properties. In this context, the low toxicity, and high pharmacokinetic properties of curcumin, which is a natural phytochemical, as well as the easy synthesizing of its derivatives reveal the need for investigation of its various derivatives as inhibitors against coronaviruses. The present study focused on curcumin derivatives with reliable ADME profile and high molecular binding potency to different SARS-CoV-2 target enzymes (3CLPro, PLpro, NSP7/8/12, NSP7/8/12 +RNA, NSP15, NSP16, Spike, Spike+ACE). In the molecular docking studies, the best binding scores for the 22 proposed curcumin derivatives were obtained for the PLpro protein. Furthermore, MD simulations were performed for high-affinity ligand-PLpro protein complexes and subsequently, Lys157, Glu161, Asp164, Arg166, Glu167, Met208, Pro247, Pro248, Tyr264, Tyr273 and Asp302 residues of PLpro was determined to play key role for ligand binding by Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) analysis. The results of the study promise that the proposed curcumin derivatives can be potent inhibitors against SARS-CoV-2 and be converted into pharmaceutical drugs. It is also expected that the findings may provide guiding insights to future design studies for synthesizing different antiviral derivatives of phytochemicals.
Keywords: Coronavirus; Curcumin; Docking; Drug design; MM-PBSA; Molecular dynamic; SARS-CoV-2; Simulation.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
-
- Adams B.K., Ferstl E.M., Davis M.C., Herold M., Kurtkaya S., Camalier R.F., Hollingshead M.G., Kaur G., Sausville E.A., Rickles F.R., Snyder J.P., Liotta D.C., Shoji M. Synthesis and biological evaluation of novel curcumin analogs as anti-cancer and anti-angiogenesis agents. Bioorg. Med. Chem. 2004;12:3871–3883. - PubMed
-
- Adelusi T.I., Oyedele A.-Q.K., Monday O.E., Boyenle I.D., Idris M.O., Ogunlana A.T., Ayoola A.M., Fatoki J.O., Kolawole O.E., David K.B., Olayemi A.A. Dietary polyphenols mitigate SARS-CoV-2 main protease (Mpro)–Molecular dynamics, molecular mechanics, and density functional theory investigations. J. Mol. Struct. 2022;1250 - PMC - PubMed
-
- Aggarwal B.B., Shishodia S., Takada Y., Banerjee S., Newman R.A., Bueso-Ramos C.E., Price J.E. Curcumin suppresses the paclitaxel-induced nuclear factor-κB pathway in breast cancer cells and inhibits lung metastasis of human breast cancer in nude mice. Clin. Cancer Res. 2005;11:7490–7498. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

