Global analysis of switchgrass (Panicum virgatum L.) transcriptomes in response to interactive effects of drought and heat stresses
- PMID: 35260072
- PMCID: PMC8903725
- DOI: 10.1186/s12870-022-03477-0
Global analysis of switchgrass (Panicum virgatum L.) transcriptomes in response to interactive effects of drought and heat stresses
Abstract
Background: Sustainable production of high-quality feedstock has been of great interest in bioenergy research. Despite the economic importance, high temperatures and water deficit are limiting factors for the successful cultivation of switchgrass in semi-arid areas. There are limited reports on the molecular basis of combined abiotic stress tolerance in switchgrass, particularly the combination of drought and heat stress. We used transcriptomic approaches to elucidate the changes in the response of switchgrass to drought and high temperature simultaneously.
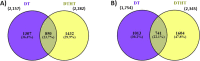
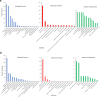
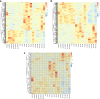
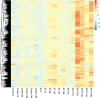
Results: We conducted solely drought treatment in switchgrass plant Alamo AP13 by withholding water after 45 days of growing. For the combination of drought and heat effect, heat treatment (35 °C/25 °C day/night) was imposed after 72 h of the initiation of drought. Samples were collected at 0 h, 72 h, 96 h, 120 h, 144 h, and 168 h after treatment imposition, total RNA was extracted, and RNA-Seq conducted. Out of a total of 32,190 genes, we identified 3912, as drought (DT) responsive genes, 2339 and 4635 as, heat (HT) and drought and heat (DTHT) responsive genes, respectively. There were 209, 106, and 220 transcription factors (TFs) differentially expressed under DT, HT and DTHT respectively. Gene ontology annotation identified the metabolic process as the significant term enriched in DTHT genes. Other biological processes identified in DTHT responsive genes included: response to water, photosynthesis, oxidation-reduction processes, and response to stress. KEGG pathway enrichment analysis on DT and DTHT responsive genes revealed that TFs and genes controlling phenylpropanoid pathways were important for individual as well as combined stress response. For example, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) from the phenylpropanoid pathway was induced by single DT and combinations of DTHT stress.
Conclusion: Through RNA-Seq analysis, we have identified unique and overlapping genes in response to DT and combined DTHT stress in switchgrass. The combination of DT and HT stress may affect the photosynthetic machinery and phenylpropanoid pathway of switchgrass which negatively impacts lignin synthesis and biomass production of switchgrass. The biological function of genes identified particularly in response to DTHT stress could further be confirmed by techniques such as single point mutation or RNAi.
Keywords: Differential gene expression; Drought stress; Gene ontology; Heat stress; Panicum virgatum; Transcription factors; Transcriptome.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Genome-wide profiling of histone (H3) lysine 4 (K4) tri-methylation (me3) under drought, heat, and combined stresses in switchgrass.BMC Genomics. 2024 Feb 29;25(1):223. doi: 10.1186/s12864-024-10068-w. BMC Genomics. 2024. PMID: 38424499 Free PMC article.
-
Transcriptome analysis of heat stress response in switchgrass (Panicum virgatum L.).BMC Plant Biol. 2013 Oct 6;13:153. doi: 10.1186/1471-2229-13-153. BMC Plant Biol. 2013. PMID: 24093800 Free PMC article.
-
Comparative transcriptome profiling of upland (VS16) and lowland (AP13) ecotypes of switchgrass.Plant Cell Rep. 2017 Jan;36(1):129-150. doi: 10.1007/s00299-016-2065-0. Epub 2016 Nov 3. Plant Cell Rep. 2017. PMID: 27812750 Free PMC article.
-
Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress.F1000Res. 2019 May 14;8:F1000 Faculty Rev-658. doi: 10.12688/f1000research.18424.1. eCollection 2019. F1000Res. 2019. PMID: 31131087 Free PMC article. Review.
-
When drought meets heat - a plant omics perspective.Front Plant Sci. 2023 Aug 22;14:1250878. doi: 10.3389/fpls.2023.1250878. eCollection 2023. Front Plant Sci. 2023. PMID: 37674736 Free PMC article. Review.
Cited by
-
Eco-Friendly Algicidal Potential of Zanthoxylum bungeanum Leaf Extracts: Extraction Optimization and Impact on Algal Growth.Microorganisms. 2025 Mar 27;13(4):760. doi: 10.3390/microorganisms13040760. Microorganisms. 2025. PMID: 40284597 Free PMC article.
-
Genome-wide analysis and identification of nuclear factor Y gene family in switchgrass (Panicum virgatum L.).BMC Genomics. 2024 Dec 20;25(1):1218. doi: 10.1186/s12864-024-11092-6. BMC Genomics. 2024. PMID: 39702036 Free PMC article.
-
Development of a Reference Transcriptome and Identification of Differentially Expressed Genes Linked to Salt Stress in Salt Marsh Grass (Sporobolus alterniflorus) along Delaware Coastal Regions.Plants (Basel). 2024 Jul 22;13(14):2008. doi: 10.3390/plants13142008. Plants (Basel). 2024. PMID: 39065534 Free PMC article.
-
GATA8-Mediated Antiviral Defence Is Countered by Tomato Chlorosis Virus-Encoded Pathogenicity Protein p27.Mol Plant Pathol. 2025 Jul;26(7):e70115. doi: 10.1111/mpp.70115. Mol Plant Pathol. 2025. PMID: 40600259 Free PMC article.
-
Ghost introgression facilitates genomic divergence of a sympatric cryptic lineage in Cycas revoluta.Ecol Evol. 2023 Aug 18;13(8):e10435. doi: 10.1002/ece3.10435. eCollection 2023 Aug. Ecol Evol. 2023. PMID: 37600490 Free PMC article.
References
-
- Sewelam N, Oshima Y, Mitsuda N, Ohme-Takagi M. A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ. 2014. 10.1111/pce.12274. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

