CMTr cap-adjacent 2'-O-ribose mRNA methyltransferases are required for reward learning and mRNA localization to synapses
- PMID: 35260552
- PMCID: PMC8904806
- DOI: 10.1038/s41467-022-28549-5
CMTr cap-adjacent 2'-O-ribose mRNA methyltransferases are required for reward learning and mRNA localization to synapses
Abstract
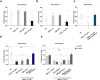
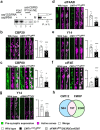
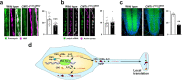
Cap-adjacent nucleotides of animal, protist and viral mRNAs can be O-methylated at the 2' position of the ribose (cOMe). The functions of cOMe in animals, however, remain largely unknown. Here we show that the two cap methyltransferases (CMTr1 and CMTr2) of Drosophila can methylate the ribose of the first nucleotide in mRNA. Double-mutant flies lack cOMe but are viable. Consistent with prominent neuronal expression, they have a reward learning defect that can be rescued by conditional expression in mushroom body neurons before training. Among CMTr targets are cell adhesion and signaling molecules. Many are relevant for learning, and are also targets of Fragile X Mental Retardation Protein (FMRP). Like FMRP, cOMe is required for localization of untranslated mRNAs to synapses and enhances binding of the cap binding complex in the nucleus. Hence, our study reveals a mechanism to co-transcriptionally prime mRNAs by cOMe for localized protein synthesis at synapses.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. - PubMed
-
- Roignant JY, Soller M. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–390. - PubMed
-
- Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020;21:36–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/L006340/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R001715/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M017982/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R002932/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

