Bacterial F-type ATP synthases follow a well-choreographed assembly pathway
- PMID: 35260553
- PMCID: PMC8904574
- DOI: 10.1038/s41467-022-28828-1
Bacterial F-type ATP synthases follow a well-choreographed assembly pathway
Abstract
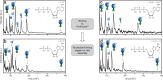
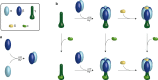
F-type ATP synthases are multiprotein complexes composed of two separate coupled motors (F1 and FO) generating adenosine triphosphate (ATP) as the universal major energy source in a variety of relevant biological processes in mitochondria, bacteria and chloroplasts. While the structure of many ATPases is solved today, the precise assembly pathway of F1FO-ATP synthases is still largely unclear. Here, we probe the assembly of the F1 complex from Acetobacterium woodii. Using laser induced liquid bead ion desorption (LILBID) mass spectrometry, we study the self-assembly of purified F1 subunits in different environments under non-denaturing conditions. We report assembly requirements and identify important assembly intermediates in vitro and in cellula. Our data provide evidence that nucleotide binding is crucial for in vitro F1 assembly, whereas ATP hydrolysis appears to be less critical. We correlate our results with activity measurements and propose a model for the assembly pathway of a functional F1 complex.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Andries K. A Diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. - PubMed
-
- De Jonge MR, Koymans LHM, Guillemont JEG, Koul A, Andries K. A computational model of the inhibition of Mycobacterium tuberculosis ATPase by a new drug candidate R207910. Proteins. 2007;67:971–980. - PubMed
-
- de Vries DD, van Engelen BGM, Gabreëls FJM, Ruitenbeek W, van Oost BA. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann. Neurol. 1993;34:410–412. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

