Positive selection over the mitochondrial genome and its role in the diversification of gentoo penguins in response to adaptation in isolation
- PMID: 35260629
- PMCID: PMC8904570
- DOI: 10.1038/s41598-022-07562-0
Positive selection over the mitochondrial genome and its role in the diversification of gentoo penguins in response to adaptation in isolation
Abstract
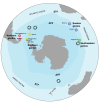
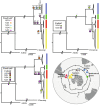
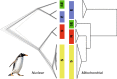
Although mitochondrial DNA has been widely used in phylogeography, evidence has emerged that factors such as climate, food availability, and environmental pressures that produce high levels of stress can exert a strong influence on mitochondrial genomes, to the point of promoting the persistence of certain genotypes in order to compensate for the metabolic requirements of the local environment. As recently discovered, the gentoo penguins (Pygoscelis papua) comprise four highly divergent lineages across their distribution spanning the Antarctic and sub-Antarctic regions. Gentoo penguins therefore represent a suitable animal model to study adaptive processes across divergent environments. Based on 62 mitogenomes that we obtained from nine locations spanning all four gentoo penguin lineages, we demonstrated lineage-specific nucleotide substitutions for various genes, but only lineage-specific amino acid replacements for the ND1 and ND5 protein-coding genes. Purifying selection (dN/dS < 1) is the main driving force in the protein-coding genes that shape the diversity of mitogenomes in gentoo penguins. Positive selection (dN/dS > 1) was mostly present in codons of the Complex I (NADH genes), supported by two different codon-based methods at the ND1 and ND4 in the most divergent lineages, the eastern gentoo penguin from Crozet and Marion Islands and the southern gentoo penguin from Antarctica respectively. Additionally, ND5 and ATP6 were under selection in the branches of the phylogeny involving all gentoo penguins except the eastern lineage. Our study suggests that local adaptation of gentoo penguins has emerged as a response to environmental variability promoting the fixation of mitochondrial haplotypes in a non-random manner. Mitogenome adaptation is thus likely to have been associated with gentoo penguin diversification across the Southern Ocean and to have promoted their survival in extreme environments such as Antarctica. Such selective processes on the mitochondrial genome may also be responsible for the discordance detected between nuclear- and mitochondrial-based phylogenies of gentoo penguin lineages.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Ballard JWO, Pichaud N. Mitochondrial DNA: More than an evolutionary bystander. Funct. Ecol. 2014;28:218–231. doi: 10.1111/1365-2435.12177. - DOI
-
- Shtolz N, Mishmar D. The mitochondrial genome–on selective constraints and signatures at the organism, cell, and single mitochondrion levels. Front. Ecol. Evol. 2019 doi: 10.3389/fevo.2019.00342. - DOI
-
- Hill GE. Mitonuclear Ecology. Oxford University Press; 2019.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

