This is a preprint.
Flipped Over U: Structural Basis for dsRNA Cleavage by the SARS-CoV-2 Endoribonuclease
- PMID: 35262076
- PMCID: PMC8902873
- DOI: 10.1101/2022.03.02.480688
Flipped Over U: Structural Basis for dsRNA Cleavage by the SARS-CoV-2 Endoribonuclease
Update in
-
Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.Nucleic Acids Res. 2022 Aug 12;50(14):8290-8301. doi: 10.1093/nar/gkac589. Nucleic Acids Res. 2022. PMID: 35801916 Free PMC article.
Abstract
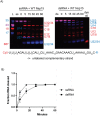
Coronaviruses generate double-stranded (ds) RNA intermediates during viral replication that can activate host immune sensors. To evade activation of the host pattern recognition receptor MDA5, coronaviruses employ Nsp15, which is uridine-specific endoribonuclease. Nsp15 is proposed to associate with the coronavirus replication-transcription complex within double-membrane vesicles to cleave these dsRNA intermediates. How Nsp15 recognizes and processes dsRNA is poorly understood because previous structural studies of Nsp15 have been limited to small single-stranded (ss) RNA substrates. Here we present cryo-EM structures of SARS-CoV-2 Nsp15 bound to a 52nt dsRNA. We observed that the Nsp15 hexamer forms a platform for engaging dsRNA across multiple protomers. The structures, along with site-directed mutagenesis and RNA cleavage assays revealed critical insight into dsRNA recognition and processing. To process dsRNA Nsp15 utilizes a base-flipping mechanism to properly orient the uridine within the active site for cleavage. Our findings show that Nsp15 is a distinctive endoribonuclease that can cleave both ss- and dsRNA effectively.
Conflict of interest statement
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
