Estimating survival parameters under conditionally independent left truncation
- PMID: 35262259
- PMCID: PMC9545094
- DOI: 10.1002/pst.2202
Estimating survival parameters under conditionally independent left truncation
Abstract
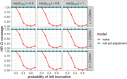
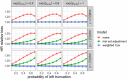
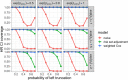
Databases derived from electronic health records (EHRs) are commonly subject to left truncation, a type of selection bias that occurs when patients need to survive long enough to satisfy certain entry criteria. Standard methods to adjust for left truncation bias rely on an assumption of marginal independence between entry and survival times, which may not always be satisfied in practice. In this work, we examine how a weaker assumption of conditional independence can result in unbiased estimation of common statistical parameters. In particular, we show the estimability of conditional parameters in a truncated dataset, and of marginal parameters that leverage reference data containing non-truncated data on confounders. The latter is complementary to observational causal inference methodology applied to real-world external comparators, which is a common use case for real-world databases. We implement our proposed methods in simulation studies, demonstrating unbiased estimation and valid statistical inference. We also illustrate estimation of a survival distribution under conditionally independent left truncation in a real-world clinico-genomic database.
Keywords: left truncation; real world data; survival analysis.
© 2022 The Author. Pharmaceutical Statistics published by John Wiley & Sons Ltd.
Conflict of interest statement
AS reports employment in Flatiron Health, which is an independent subsidiary of the Roche group and stock ownership in Roche.
Figures



Similar articles
-
Penalized regression for left-truncated and right-censored survival data.Stat Med. 2021 Nov 10;40(25):5487-5500. doi: 10.1002/sim.9136. Epub 2021 Jul 24. Stat Med. 2021. PMID: 34302373 Free PMC article.
-
Cox regression model under dependent truncation.Biometrics. 2022 Jun;78(2):460-473. doi: 10.1111/biom.13451. Epub 2021 Mar 22. Biometrics. 2022. PMID: 33687064 Free PMC article.
-
Nonparametric estimation of the survival distribution under covariate-induced dependent truncation.Biometrics. 2022 Dec;78(4):1390-1401. doi: 10.1111/biom.13545. Epub 2021 Aug 22. Biometrics. 2022. PMID: 34389985
-
Insights into the Cross-world Independence Assumption of Causal Mediation Analysis.Epidemiology. 2021 Mar 1;32(2):209-219. doi: 10.1097/EDE.0000000000001313. Epidemiology. 2021. PMID: 33512846 Review.
-
Estimating a time-to-event distribution from right-truncated data in an epidemic: A review of methods.Stat Methods Med Res. 2022 Sep;31(9):1641-1655. doi: 10.1177/09622802211023955. Epub 2021 Dec 21. Stat Methods Med Res. 2022. PMID: 34931911 Free PMC article. Review.
Cited by
-
Cancer survival: left truncation and comparison of results from hospital-based cancer registry and population-based cancer registry.Front Oncol. 2023 Jun 7;13:1173828. doi: 10.3389/fonc.2023.1173828. eCollection 2023. Front Oncol. 2023. PMID: 37350938 Free PMC article.
-
Elucidating Analytic Bias Due to Informative Cohort Entry in Cancer Clinico-genomic Datasets.Cancer Epidemiol Biomarkers Prev. 2023 Mar 6;32(3):344-352. doi: 10.1158/1055-9965.EPI-22-0875. Cancer Epidemiol Biomarkers Prev. 2023. PMID: 36626408 Free PMC article.
-
Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning.Cancers (Basel). 2023 Mar 20;15(6):1853. doi: 10.3390/cancers15061853. Cancers (Basel). 2023. PMID: 36980739 Free PMC article.
-
Brief Report: Not Created Equal: Survival Differences by KRAS Mutation Subtype in NSCLC Treated With Immunotherapy.JTO Clin Res Rep. 2024 Oct 24;6(1):100755. doi: 10.1016/j.jtocrr.2024.100755. eCollection 2025 Jan. JTO Clin Res Rep. 2024. PMID: 39758602 Free PMC article.
-
Biomarker Inference and the Timing of Next-Generation Sequencing in a Multi-Institutional, Cross-Cancer Clinicogenomic Data Set.JCO Precis Oncol. 2024 Mar;8:e2300489. doi: 10.1200/PO.23.00489. JCO Precis Oncol. 2024. PMID: 38484212 Free PMC article.
References
-
- Klein JP, Moeschberger ML. Survival Analysis: Techniques for Censored and Truncated Data. Vol 1230. Springer; 2003.
-
- Agarwala V, Khozin S, Singal G, et al. Real‐world evidence in support of precision medicine: clinico‐genomic cancer data as a case study. Health Aff. 2018;37(5):765‐772. - PubMed
-
- Tsai WY, Jewell NP, Wang MC. A note on the product‐limit estimator under right censoring and left truncation. Biometrika. 1987;74(4):883‐886.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

