Tumor cell intrinsic and extrinsic features predict prognosis in estrogen receptor positive breast cancer
- PMID: 35263321
- PMCID: PMC8936467
- DOI: 10.1371/journal.pcbi.1009495
Tumor cell intrinsic and extrinsic features predict prognosis in estrogen receptor positive breast cancer
Abstract
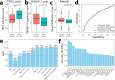
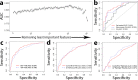
Although estrogen-receptor-positive (ER+) breast cancer is generally associated with favorable prognosis, clinical outcome varies substantially among patients. Genomic assays have been developed and applied to predict patient prognosis for personalized treatment. We hypothesize that the recurrence risk of ER+ breast cancer patients is determined by both genomic mutations intrinsic to tumor cells and extrinsic immunological features in the tumor microenvironment. Based on the Cancer Genome Atlas (TCGA) breast cancer data, we identified the 72 most common genomic aberrations (including gene mutations and indels) in ER+ breast cancer and defined sample-specific scores that systematically characterized the deregulated pathways intrinsic to tumor cells. To further consider tumor cell extrinsic features, we calculated immune infiltration scores for six major immune cell types. Many individual intrinsic features are predictive of patient prognosis in ER+ breast cancer, and some of them achieved comparable accuracy with the Oncotype DX assay. In addition, statistical learning models that integrated these features predicts the recurrence risk of patients with significantly better performance than the Oncotype DX assay (our optimized random forest model AUC = 0.841, Oncotype DX model AUC = 0.792, p = 0.04). As a proof-of-concept, our study indicates the great potential of genomic and immunological features in prognostic prediction for improving breast cancer precision medicine. The framework introduced in this work can be readily applied to other cancers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Relationship of Oncotype Dx score with tumor grade, size, nodal status, proliferative marker Ki67 and Nottingham Prognostic Index in early breast cancer tumors in Saudi Population.Ann Diagn Pathol. 2021 Apr;51:151674. doi: 10.1016/j.anndiagpath.2020.151674. Epub 2020 Nov 25. Ann Diagn Pathol. 2021. PMID: 33360027
-
Oncotype DX® in breast cancer patients: clinical experience, outcome and follow-up-a case-control study.Arch Gynecol Obstet. 2018 Feb;297(2):443-447. doi: 10.1007/s00404-017-4618-z. Epub 2017 Dec 13. Arch Gynecol Obstet. 2018. PMID: 29236174
-
Is oncotype DX recurrence score (RS) of prognostic value once HER2-positive and. low-ER expression patients are removed?Breast J. 2013 Jul-Aug;19(4):357-64. doi: 10.1111/tbj.12126. Epub 2013 May 23. Breast J. 2013. PMID: 23701403
-
Prognostic and Predictive Biomarkers of Endocrine Responsiveness for Estrogen Receptor Positive Breast Cancer.Adv Exp Med Biol. 2016;882:125-54. doi: 10.1007/978-3-319-22909-6_5. Adv Exp Med Biol. 2016. PMID: 26987533 Review.
-
Multigene prognostic tests in breast cancer: past, present, future.Breast Cancer Res. 2015 Jan 27;17(1):11. doi: 10.1186/s13058-015-0514-2. Breast Cancer Res. 2015. PMID: 25848861 Free PMC article. Review.
References
-
- Henry N, Shah P, Haider I, Freer P, Jagsi R, Sabel M. Chapter 88: cancer of the breast. Abeloff’s clinical oncology, 6th edn Elsevier, Philadelphia, PA. 2020.
-
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al.. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor–positive breast cancer. Clinical cancer research. 2010;16(21):5222–32. doi: 10.1158/1078-0432.CCR-10-1282 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

