A nonredundant role for T cell-derived interleukin 22 in antibacterial defense of colonic crypts
- PMID: 35263568
- PMCID: PMC9126440
- DOI: 10.1016/j.immuni.2022.02.003
A nonredundant role for T cell-derived interleukin 22 in antibacterial defense of colonic crypts
Abstract
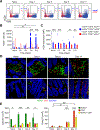
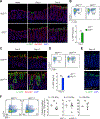
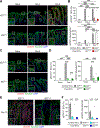
Interleukin (IL)-22 is central to immune defense at barrier sites. We examined the contributions of innate lymphoid cell (ILC) and T cell-derived IL-22 during Citrobacter rodentium (C.r) infection using mice that both report Il22 expression and allow lineage-specific deletion. ILC-derived IL-22 activated STAT3 in C.r-colonized surface intestinal epithelial cells (IECs) but only temporally restrained bacterial growth. T cell-derived IL-22 induced a more robust and extensive activation of STAT3 in IECs, including IECs lining colonic crypts, and T cell-specific deficiency of IL-22 led to pathogen invasion of the crypts and increased mortality. This reflected a requirement for T cell-derived IL-22 for the expression of a host-protective transcriptomic program that included AMPs, neutrophil-recruiting chemokines, and mucin-related molecules, and it restricted IFNγ-induced proinflammatory genes. Our findings demonstrate spatiotemporal differences in the production and action of IL-22 by ILCs and T cells during infection and reveal an indispensable role for IL-22-producing T cells in the protection of the intestinal crypts.
Keywords: AMPs; CD4 T cells; Citrobacter rodentium; IFNγ; IL-22; TNF; chemokines; colonic crypt IECs; colonic surface IECs; innate cells; mucins.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Akdis M, Palomares O, van de Veen W, van Splunter M, and Akdis CA (2012). TH17 and TH22 cells: A confusion of antimicrobial response with tissue inflammation versus protection. J Allergy Clin Immun 129, 1438–1449. - PubMed
-
- Allen A, Hutton DA, and Pearson JP (1998). The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biology 30, 797–801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

