Mechanistic convergence of the TIGIT and PD-1 inhibitory pathways necessitates co-blockade to optimize anti-tumor CD8+ T cell responses
- PMID: 35263569
- PMCID: PMC9287124
- DOI: 10.1016/j.immuni.2022.02.005
Mechanistic convergence of the TIGIT and PD-1 inhibitory pathways necessitates co-blockade to optimize anti-tumor CD8+ T cell responses
Abstract
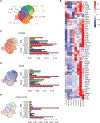
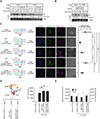
Dual blockade of the PD-1 and TIGIT coinhibitory receptors on T cells shows promising early results in cancer patients. Here, we studied the mechanisms whereby PD-1 and/or TIGIT blockade modulate anti-tumor CD8+ T cells. Although PD-1 and TIGIT are thought to regulate different costimulatory receptors (CD28 and CD226), effectiveness of PD-1 or TIGIT inhibition in preclinical tumor models was reduced in the absence of CD226. CD226 expression associated with clinical benefit in patients with non-small cell lung carcinoma (NSCLC) treated with anti-PD-L1 antibody atezolizumab. CD226 and CD28 were co-expressed on NSCLC infiltrating CD8+ T cells poised for expansion. Mechanistically, PD-1 inhibited phosphorylation of both CD226 and CD28 via its ITIM-containing intracellular domain (ICD); TIGIT's ICD was dispensable, with TIGIT restricting CD226 co-stimulation by blocking interaction with their common ligand PVR (CD155). Thus, full restoration of CD226 signaling, and optimal anti-tumor CD8+ T cell responses, requires blockade of TIGIT and PD-1, providing a mechanistic rationale for combinatorial targeting in the clinic.
Keywords: CD226; cancer immunotherapy; costimulatory receptor TIGIT; immune checkpoint blockade.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.L.B., A.S.C., A.A.-Y., W.E.O., T.D.W., S.M., R.C., L.C.-A., J.L.G., I.M., and E.Y.C. are or were employees of Genentech, a member of the Roche group, which develops and markets drugs for profit.
Figures




References
-
- Amsen D, van Gisbergen KPJM, Hombrink P, and van Lier RAW (2018). Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 19, 538–546. - PubMed
-
- Braun M, Aguilera AR, Sundarrajan A, Corvino D, Stannard K, Krumeich S, Das I, Lima LG, Meza Guzman LG, Li K, et al. (2020). CD155 on Tumor Cells Drives Resistance to Immunotherapy by Inducing the Degradation of the Activating Receptor CD226 in CD8+ T Cells. Immunity 53, 805–823.e15. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

