Visualization of the process of a nanocarrier-mediated gene delivery: stabilization, endocytosis and endosomal escape of genes for intracellular spreading
- PMID: 35264206
- PMCID: PMC8905852
- DOI: 10.1186/s12951-022-01336-6
Visualization of the process of a nanocarrier-mediated gene delivery: stabilization, endocytosis and endosomal escape of genes for intracellular spreading
Abstract
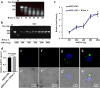
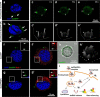
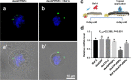
Nanoparticles have been widely applied as gene carrier for improving RNA interference (RNAi) efficiency in medical and agricultural fields. However, the mechanism and delivery process of nanoparticle-mediated RNAi is not directly visualized and elucidated. Here we synthesized a star polymer (SPc) consisted of a hydrophilic shell with positively-charged tertiary amine in the side chain, which was taken as an example to investigate the mechanism in gene delivery. The SPc could assemble with dsRNA spontaneously through electrostatic force, hydrogen bond and van der Waals force. Interestingly, the SPc could protect dsRNA from degradation by RNase A and insect hemolymph, thus remarkably increasing the stability of dsRNA. Meanwhile, the SPc could efficiently promote the cellular uptake and endosomal escape for intracellular spreading of dsRNA. Transcriptome analysis revealed that the SPc could up-regulate some key genes such as Chc, AP2S1 and Arf1 for activating clathrin-mediated endocytosis. Furthermore, the suppression of endocytosis hindered the cellular uptake of SPc-delivered dsRNA in vitro, and the subsequent RNAi effect was also disappeared in vivo. To our knowledge, our study is the first direct visualization of the detailed cellular delivery process and mechanism of nanocarrier-mediated gene delivery. Above mechanism supports the application of nanocarrier-based RNAi in gene therapy and pest management.
Keywords: Cellular uptake; Clathrin; Endocytosis; Nanocarrier; RNA interference; dsRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Nano-delivery platform with strong protection and efficient delivery: preparation of self-assembled RNA pesticide with dual RNAi targets against Apolygus lucorum.J Nanobiotechnology. 2025 Feb 7;23(1):93. doi: 10.1186/s12951-025-03155-x. J Nanobiotechnology. 2025. PMID: 39920702 Free PMC article.
-
Chitosan/dsRNA polyplex nanoparticles advance environmental RNA interference efficiency through activating clathrin-dependent endocytosis.Int J Biol Macromol. 2023 Dec 31;253(Pt 4):127021. doi: 10.1016/j.ijbiomac.2023.127021. Epub 2023 Sep 21. Int J Biol Macromol. 2023. PMID: 37741481
-
Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle.Insect Biochem Mol Biol. 2015 May;60:68-77. doi: 10.1016/j.ibmb.2015.03.009. Epub 2015 Apr 9. Insect Biochem Mol Biol. 2015. PMID: 25863352
-
Nanoparticle-mediated double-stranded RNA delivery system: A promising approach for sustainable pest management.Insect Sci. 2021 Feb;28(1):21-34. doi: 10.1111/1744-7917.12822. Epub 2020 Jul 20. Insect Sci. 2021. PMID: 32478473 Review.
-
The mysteries of insect RNAi: A focus on dsRNA uptake and transport.Pestic Biochem Physiol. 2018 Oct;151:25-31. doi: 10.1016/j.pestbp.2018.08.005. Epub 2018 Aug 17. Pestic Biochem Physiol. 2018. PMID: 30704709 Review.
Cited by
-
Recent Advances in Nanoparticle-Mediated Co-Delivery System: A Promising Strategy in Medical and Agricultural Field.Int J Mol Sci. 2023 Mar 7;24(6):5121. doi: 10.3390/ijms24065121. Int J Mol Sci. 2023. PMID: 36982200 Free PMC article. Review.
-
The role of polymers in enabling RNAi-based technology for sustainable pest management.Nat Commun. 2024 Oct 23;15(1):9158. doi: 10.1038/s41467-024-53468-y. Nat Commun. 2024. PMID: 39443470 Free PMC article. Review.
-
Chronic exposure to the star polycation (SPc) nanocarrier in the larval stage adversely impairs life history traits in Drosophila melanogaster.J Nanobiotechnology. 2022 Dec 8;20(1):515. doi: 10.1186/s12951-022-01705-1. J Nanobiotechnology. 2022. PMID: 36482441 Free PMC article.
-
Calcium nutrition nanoagent rescues tomatoes from mosaic virus disease by accelerating calcium transport and activating antiviral immunity.Front Plant Sci. 2022 Dec 6;13:1092774. doi: 10.3389/fpls.2022.1092774. eCollection 2022. Front Plant Sci. 2022. PMID: 36561462 Free PMC article.
-
RNA Interference in Insects: From a Natural Mechanism of Gene Expression Regulation to a Biotechnological Crop Protection Promise.Biology (Basel). 2024 Feb 21;13(3):137. doi: 10.3390/biology13030137. Biology (Basel). 2024. PMID: 38534407 Free PMC article. Review.
References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Van Rij RP, Andino R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–193. - PubMed
-
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. - PubMed
-
- Saw PE, Song EW. siRNA therapeutics: A clinical reality. Sci China Life Sci. 2020;63:485–500. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

