A prospective observational cohort study to identify inflammatory biomarkers for the diagnosis and prognosis of patients with sepsis
- PMID: 35264246
- PMCID: PMC8905560
- DOI: 10.1186/s40560-022-00602-x
A prospective observational cohort study to identify inflammatory biomarkers for the diagnosis and prognosis of patients with sepsis
Abstract
Background: Sepsis is a life-threatening organ dysfunction. A fast diagnosis is crucial for patient management. Proteins that are synthesized during the inflammatory response can be used as biomarkers, helping in a rapid clinical assessment or an early diagnosis of infection. The aim of this study was to identify biomarkers of inflammation for the diagnosis and prognosis of infection in patients with suspected sepsis.
Methods: In total 406 episodes were included in a prospective cohort study. Plasma was collected from all patients with suspected sepsis, for whom blood cultures were drawn, in the emergency department (ED), the department of infectious diseases, or the haemodialysis unit on the first day of a new episode. Samples were analysed using a 92-plex proteomic panel based on a proximity extension assay with oligonucleotide-labelled antibody probe pairs (OLink, Uppsala, Sweden). Supervised and unsupervised differential expression analyses and pathway enrichment analyses were performed to search for inflammatory proteins that were different between patients with viral or bacterial sepsis and between patients with worse or less severe outcome.
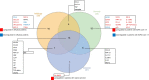
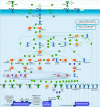
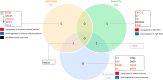
Results: Supervised differential expression analysis revealed 21 proteins that were significantly lower in circulation of patients with viral infections compared to patients with bacterial infections. More strongly, higher expression levels were observed for 38 proteins in patients with high SOFA scores (> 4), and for 21 proteins in patients with worse outcome. These proteins are mostly involved in pathways known to be activated early in the inflammatory response. Unsupervised, hierarchical clustering confirmed that inflammatory response was more strongly related to disease severity than to aetiology.
Conclusion: Several differentially expressed inflammatory proteins were identified that could be used as biomarkers for sepsis. These proteins are mostly related to disease severity. Within the setting of an emergency department, they could be used for outcome prediction, patient monitoring, and directing diagnostics.
Trail registration number: clinicaltrial.gov identifier NCT03841162.
Keywords: Biomarkers; Disease severity; Inflammation; Sepsis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
A hypolipoprotein sepsis phenotype indicates reduced lipoprotein antioxidant capacity, increased endothelial dysfunction and organ failure, and worse clinical outcomes.Crit Care. 2021 Sep 17;25(1):341. doi: 10.1186/s13054-021-03757-5. Crit Care. 2021. PMID: 34535154 Free PMC article.
-
Sepsis Care Pathway 2019.Qatar Med J. 2019 Nov 7;2019(2):4. doi: 10.5339/qmj.2019.qccc.4. eCollection 2019. Qatar Med J. 2019. PMID: 31763206 Free PMC article.
-
Clinical evaluation of droplet digital PCR in the early identification of suspected sepsis patients in the emergency department: a prospective observational study.Front Cell Infect Microbiol. 2024 Jun 4;14:1358801. doi: 10.3389/fcimb.2024.1358801. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38895732 Free PMC article.
-
Prospective validation of a transcriptomic severity classifier among patients with suspected acute infection and sepsis in the emergency department.Eur J Emerg Med. 2022 Oct 1;29(5):357-365. doi: 10.1097/MEJ.0000000000000931. Epub 2022 Apr 21. Eur J Emerg Med. 2022. PMID: 35467566 Free PMC article.
-
Inflammatory biomarkers to predict the prognosis of acute bacterial and viral infections.J Crit Care. 2023 Dec;78:154360. doi: 10.1016/j.jcrc.2023.154360. Epub 2023 Jun 19. J Crit Care. 2023. PMID: 37343422 Review.
Cited by
-
Identification of biomarkers and therapeutic targets related to Sepsis-associated encephalopathy in rats by quantitative proteomics.BMC Genomics. 2023 Jan 4;24(1):4. doi: 10.1186/s12864-022-09101-7. BMC Genomics. 2023. PMID: 36600206 Free PMC article.
-
A cytokine/PTX3 prognostic index as a predictor of mortality in sepsis.Front Immunol. 2022 Sep 15;13:979232. doi: 10.3389/fimmu.2022.979232. eCollection 2022. Front Immunol. 2022. PMID: 36189302 Free PMC article.
-
Rapid and Differential Diagnosis of Sepsis Stages Using an Advanced 3D Plasmonic Bimetallic Alloy Nanoarchitecture-Based SERS Biosensor Combined with Machine Learning for Multiple Analyte Identification.Adv Sci (Weinh). 2025 Apr;12(14):e2414688. doi: 10.1002/advs.202414688. Epub 2025 Feb 17. Adv Sci (Weinh). 2025. PMID: 39960361 Free PMC article.
-
Synthetic plasma pool cohort correction for affinity-based proteomics datasets allows multiple study comparison.Brief Bioinform. 2024 Nov 22;26(1):bbae657. doi: 10.1093/bib/bbae657. Brief Bioinform. 2024. PMID: 39694815 Free PMC article.
-
Performance effectiveness of vital parameter combinations for early warning of sepsis-an exhaustive study using machine learning.JAMIA Open. 2022 Oct 14;5(4):ooac080. doi: 10.1093/jamiaopen/ooac080. eCollection 2022 Dec. JAMIA Open. 2022. PMID: 36267121 Free PMC article.
References
-
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. - DOI - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

