Genomic patterns of homozygosity and inbreeding depression in Murciano-Granadina goats
- PMID: 35264251
- PMCID: PMC8908635
- DOI: 10.1186/s40104-022-00684-5
Genomic patterns of homozygosity and inbreeding depression in Murciano-Granadina goats
Abstract
Background: Inbreeding depression can adversely affect traits related to fitness, reproduction and productive performance. Although current research suggests that inbreeding levels are generally low in most goat breeds, the impact of inbreeding depression on phenotypes of economic interest has only been investigated in a few studies based on genealogical data.
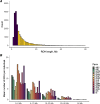
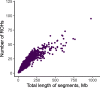
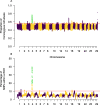
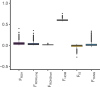
Results: We genotyped 1040 goats with the Goat SNP50 BeadChip. This information was used to estimate different molecular inbreeding coefficients and characterise runs of homozygosity and homozygosity patterns. We detected 38 genomic regions with increased homozygosity as well as 8 ROH hotspots mapping to chromosomes 1, 2, 4, 6, 14, 16 and 17. Eight hundred seventeen goats with available records for dairy traits were analysed to evaluate the potential consequences of inbreeding depression on milk phenotypes. Four regions on chromosomes 8 and 25 were significantly associated with inbreeding depression for the natural logarithm of the somatic cell count. Notably, these regions contain several genes related with immunity, such as SYK, IL27, CCL19 and CCL21. Moreover, one region on chromosome 2 was significantly associated with inbreeding depression for milk yield.
Conclusions: Although genomic inbreeding levels are low in Murciano-Granadina goats, significant evidence of inbreeding depression for the logarithm of the somatic cell count, a phenotype closely associated with udder health and milk yield, have been detected in this population. Minimising inbreeding would be expected to augment economic gain by increasing milk yield and reducing the incidence of mastitis, which is one of the main causes of dairy goat culling.
Keywords: Goat; Inbreeding; Milk yield; Murciano-Granadina; Somatic cell score.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Lush JL. The consequences and measurement of inbreeding. In: Animal breeding plans. Ames: The Iowa State College Press; 1943. 259–288.
-
- McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, MacLeod AK, Farrington SM, Rudan P, Hayward C, Vitart V, Rudan I, Wild SH, Dunlop MG, Wright AF, Campbell H, Wilson JF. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83(3):359–372. doi: 10.1016/j.ajhg.2008.08.007. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

