GC content, but not nucleosome positioning, directly contributes to intron splicing efficiency in Paramecium
- PMID: 35264448
- PMCID: PMC8997360
- DOI: 10.1101/gr.276125.121
GC content, but not nucleosome positioning, directly contributes to intron splicing efficiency in Paramecium
Abstract
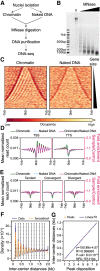
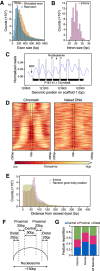
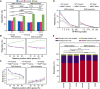
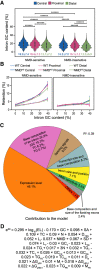
Eukaryotic genes are interrupted by introns that must be accurately spliced from mRNA precursors. With an average length of 25 nt, the more than 90,000 introns of Paramecium tetraurelia stand among the shortest introns reported in eukaryotes. The mechanisms specifying the correct recognition of these tiny introns remain poorly understood. Splicing can occur cotranscriptionally, and it has been proposed that chromatin structure might influence splice site recognition. To investigate the roles of nucleosome positioning in intron recognition, we determined the nucleosome occupancy along the P. tetraurelia genome. We show that P. tetraurelia displays a regular nucleosome array with a nucleosome repeat length of ∼151 bp, among the smallest periodicities reported. Our analysis has revealed that introns are frequently associated with inter-nucleosomal DNA, pointing to an evolutionary constraint favoring introns at the AT-rich nucleosome edge sequences. Using accurate splicing efficiency data from cells depleted for nonsense-mediated decay effectors, we show that introns located at the edge of nucleosomes display higher splicing efficiency than those at the center. However, multiple regression analysis indicates that the low GC content of introns, rather than nucleosome positioning, is associated with high splicing efficiency. Our data reveal a complex link between GC content, nucleosome positioning, and intron evolution in Paramecium.
© 2022 Gnan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
