CRISPR/Cas9-Mediated Insertion of HIV Long Terminal Repeat within BACH2 Promotes Expansion of T Regulatory-like Cells
- PMID: 35264460
- PMCID: PMC8976747
- DOI: 10.4049/jimmunol.2100491
CRISPR/Cas9-Mediated Insertion of HIV Long Terminal Repeat within BACH2 Promotes Expansion of T Regulatory-like Cells
Abstract
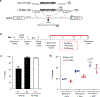
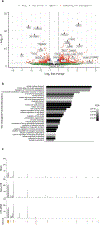
One key barrier to curative therapies for HIV is the limited understanding of HIV persistence. HIV provirus integration sites (ISs) within BACH2 are common, and almost all sites mapped to date are located upstream of the start codon in the same transcriptional orientation as the gene. These unique features suggest the possibility of insertional mutagenesis at this location. Using CRISPR/Cas9-based homology-directed repair in primary human CD4+ T cells, we directly modeled the effects of HIV integration within BACH2 Integration of the HIV long terminal repeat (LTR) and major splice donor increased BACH2 mRNA and protein levels, altered gene expression, and promoted selective outgrowth of an activated, proliferative, and T regulatory-like cell population. In contrast, introduction of the HIV-LTR alone or an HIV-LTR-major splice donor construct into STAT5B, a second common HIV IS, had no functional impact. Thus, HIV LTR-driven BACH2 expression modulates T cell programming and leads to cellular outgrowth and unique phenotypic changes, findings that support a direct role for IS-dependent HIV-1 persistence.
Copyright © 2022 by The American Association of Immunologists, Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



References
-
- Schröder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. 2002. HIV-1 Integration in the Human Genome Favors Active Genes and Local Hotspots. Cell. 110: 521–529. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

