A large and diverse autosomal haplotype is associated with sex-linked colour polymorphism in the guppy
- PMID: 35264556
- PMCID: PMC8907176
- DOI: 10.1038/s41467-022-28895-4
A large and diverse autosomal haplotype is associated with sex-linked colour polymorphism in the guppy
Abstract
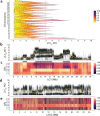
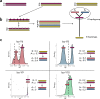
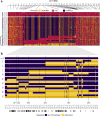
Male colour patterns of the Trinidadian guppy (Poecilia reticulata) are typified by extreme variation governed by both natural and sexual selection. Since guppy colour patterns are often inherited faithfully from fathers to sons, it has been hypothesised that many of the colour trait genes must be physically linked to sex determining loci as a 'supergene' on the sex chromosome. Here, we phenotype and genotype four guppy 'Iso-Y lines', where colour was inherited along the patriline for 40 generations. Using an unbiased phenotyping method, we confirm the breeding design was successful in creating four distinct colour patterns. We find that genetic differentiation among the Iso-Y lines is repeatedly associated with a diverse haplotype on an autosome (LG1), not the sex chromosome (LG12). Moreover, the LG1 haplotype exhibits elevated linkage disequilibrium and evidence of sex-specific diversity in the natural source population. We hypothesise that colour pattern polymorphism is driven by Y-autosome epistasis.
© 2022. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 2009;10:797–804. - PubMed
-
- Mckinnon JS, Pierotti MER. Colour polymorphism and correlated characters: genetic mechanisms and evolution. Mol. Ecol. 2010;19:5101–5125. - PubMed
-
- Svensson EI. Back to basics: using colour polymorphisms to study evolutionary processes. Mol. Ecol. 2017;26:2204–2211. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

