Elucidating mechanisms of genetic cross-disease associations at the PROCR vascular disease locus
- PMID: 35264566
- PMCID: PMC8907312
- DOI: 10.1038/s41467-022-28729-3
Elucidating mechanisms of genetic cross-disease associations at the PROCR vascular disease locus
Erratum in
-
Publisher Correction: Elucidating mechanisms of genetic cross-disease associations at the PROCR vascular disease locus.Nat Commun. 2022 Apr 6;13(1):1962. doi: 10.1038/s41467-022-29641-6. Nat Commun. 2022. PMID: 35388009 Free PMC article. No abstract available.
Abstract
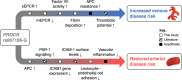
Many individual genetic risk loci have been associated with multiple common human diseases. However, the molecular basis of this pleiotropy often remains unclear. We present an integrative approach to reveal the molecular mechanism underlying the PROCR locus, associated with lower coronary artery disease (CAD) risk but higher venous thromboembolism (VTE) risk. We identify PROCR-p.Ser219Gly as the likely causal variant at the locus and protein C as a causal factor. Using genetic analyses, human recall-by-genotype and in vitro experimentation, we demonstrate that PROCR-219Gly increases plasma levels of (activated) protein C through endothelial protein C receptor (EPCR) ectodomain shedding in endothelial cells, attenuating leukocyte-endothelial cell adhesion and vascular inflammation. We also associate PROCR-219Gly with an increased pro-thrombotic state via coagulation factor VII, a ligand of EPCR. Our study, which links PROCR-219Gly to CAD through anti-inflammatory mechanisms and to VTE through pro-thrombotic mechanisms, provides a framework to reveal the mechanisms underlying similar cross-phenotype associations.
© 2022. The Author(s).
Conflict of interest statement
During the drafting of the manuscript, L.C. and J.M.M.H. became full-time employees of Novo Nordisk and D.S.P. became a full-time employee of AstraZeneca. J.E.P. has received travel and accommodation expenses and hospitality from Olink to speak at Olink-sponsored academic meetings. A.D.G., D.J.S. and N.J. are employees and stakeholders of SomaLogic. J.D. reports grants, personal fees and non-financial support from Merck Sharp & Dohme (MSD); grants, personal fees and non-financial support from Novartis; grants from Pfizer; and grants from AstraZeneca outside the submitted work. J.D. sits on the International Cardiovascular and Metabolic Advisory Board for Novartis (since 2010); the Steering Committee of UK Biobank (since 2011); the MRC International Advisory Group (ING) member, London (since 2013); the MRC High Throughput Science ‘Omics Panel Member, London (since 2013); the Scientific Advisory Committee for Sanofi (since 2013); the International Cardiovascular and Metabolism Research and Development Portfolio Committee for Novartis; and the AstraZeneca Genomics Advisory Board (2018). The remaining authors declare no competing interests.
Figures





References
-
- Medina P, et al. Endothelial protein C receptor polymorphisms and risk of myocardial infarction. Haematologica. 2008;93:1358–1363. - PubMed
MeSH terms
Substances
Grants and funding
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- BRC-1215-20014/DH_/Department of Health/United Kingdom
- RE/13/6/30180/BHF_/British Heart Foundation/United Kingdom
- MR/P502091/1/MRC_/Medical Research Council/United Kingdom
- UL1 RR025005/RR/NCRR NIH HHS/United States
- NIHR133788/DH_/Department of Health/United Kingdom
- R01 HL086694/HL/NHLBI NIH HHS/United States
- RG/18/13/33946/BHF_/British Heart Foundation/United Kingdom
- RG/19/9/34655/BHF_/British Heart Foundation/United Kingdom
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- RG/16/4/32218/BHF_/British Heart Foundation/United Kingdom
- 204623/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- R01 HL134894/HL/NHLBI NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- R01 HL059367/HL/NHLBI NIH HHS/United States
- MR/S004068/2/MRC_/Medical Research Council/United Kingdom
- U01 HG004402/HG/NHGRI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- MC_UU_00002/7/MRC_/Medical Research Council/United Kingdom
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- CSO_/Chief Scientist Office/United Kingdom
- R01 HL087641/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- RG/13/13/30194/BHF_/British Heart Foundation/United Kingdom
- SP/19/2/344612/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

