The flavonoid corylin exhibits lifespan extension properties in mouse
- PMID: 35264584
- PMCID: PMC8907184
- DOI: 10.1038/s41467-022-28908-2
The flavonoid corylin exhibits lifespan extension properties in mouse
Abstract
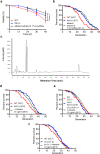
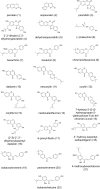
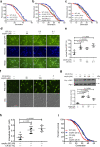
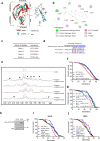
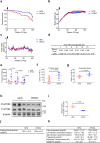
In the long history of traditional Chinese medicine, single herbs and complex formulas have been suggested to increase lifespan. However, the identification of single molecules responsible for lifespan extension has been challenging. Here, we collected a list of traditional Chinese medicines with potential longevity properties from pharmacopeias. By utilizing the mother enrichment program, we systematically screened these traditional Chinese medicines and identified a single herb, Psoralea corylifolia, that increases lifespan in Saccharomyces cerevisiae. Next, twenty-two pure compounds were isolated from Psoralea corylifolia. One of the compounds, corylin, was found to extend the replicative lifespan in yeast by targeting the Gtr1 protein. In human umbilical vein endothelial cells, RNA sequencing data showed that corylin ameliorates cellular senescence. We also examined an in vivo mammalian model, and found that corylin extends lifespan in mice fed a high-fat diet. Taken together, these findings suggest that corylin may promote longevity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

